
Bovine faeces associated smacovirus 3
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cremevirales; Smacoviridae; Bovismacovirus; Bovismacovirus bovas2
Average proteome isoelectric point is 5.89
Get precalculated fractions of proteins
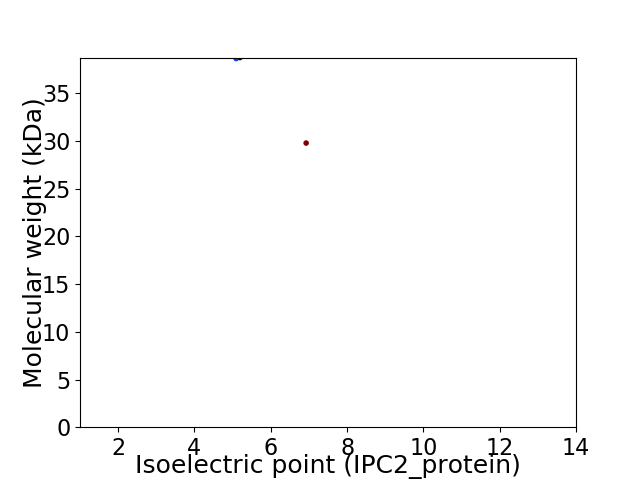
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A161IB94|A0A161IB94_9VIRU Putative capsid protein OS=Bovine faeces associated smacovirus 3 OX=1843751 PE=4 SV=1
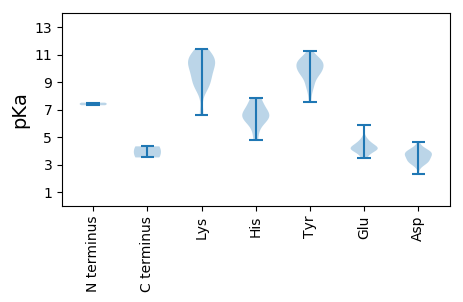
MM1 pKa = 7.31EE2 pKa = 5.87AFRR5 pKa = 11.84MVSVTLRR12 pKa = 11.84EE13 pKa = 4.16TYY15 pKa = 10.57DD16 pKa = 3.5LSTKK20 pKa = 8.55QGKK23 pKa = 6.68MTLIGIHH30 pKa = 6.54IPTSNIIQRR39 pKa = 11.84LYY41 pKa = 10.48PGFCEE46 pKa = 3.99QYY48 pKa = 9.9KK49 pKa = 8.73MCRR52 pKa = 11.84INYY55 pKa = 7.55QNVRR59 pKa = 11.84IEE61 pKa = 4.26SASRR65 pKa = 11.84LPLDD69 pKa = 3.71PAGVGVNEE77 pKa = 4.54EE78 pKa = 4.42GLVSPEE84 pKa = 4.31DD85 pKa = 3.56VFNPILYY92 pKa = 9.92KK93 pKa = 10.64AVSNQSLSNLEE104 pKa = 3.55YY105 pKa = 10.48RR106 pKa = 11.84IRR108 pKa = 11.84GLRR111 pKa = 11.84DD112 pKa = 2.94QEE114 pKa = 4.25LSVDD118 pKa = 4.07GEE120 pKa = 4.47SATASNEE127 pKa = 3.75NVTFLDD133 pKa = 3.92DD134 pKa = 3.93EE135 pKa = 5.03FNVYY139 pKa = 10.57YY140 pKa = 10.7SILSNTRR147 pKa = 11.84GWKK150 pKa = 8.69VAHH153 pKa = 6.23PQAGLSMRR161 pKa = 11.84KK162 pKa = 9.14LRR164 pKa = 11.84PLVFEE169 pKa = 4.15KK170 pKa = 10.3WFSEE174 pKa = 4.24GVNGVNDD181 pKa = 3.18FTIPVDD187 pKa = 3.73SSHH190 pKa = 7.7LEE192 pKa = 3.91GEE194 pKa = 4.5TPVNYY199 pKa = 9.4IGQMPDD205 pKa = 2.29EE206 pKa = 4.6AMRR209 pKa = 11.84GRR211 pKa = 11.84PHH213 pKa = 7.84AMPAFNTKK221 pKa = 10.23YY222 pKa = 9.76LTGVQDD228 pKa = 3.5GSGAGVGKK236 pKa = 9.37FQQNGMGDD244 pKa = 4.18GLPGNAQVSMPILPTIYY261 pKa = 9.6TGLILVPPSRR271 pKa = 11.84NTTLYY276 pKa = 9.58YY277 pKa = 10.66RR278 pKa = 11.84MVVEE282 pKa = 4.47TVVSFWGVQTMDD294 pKa = 4.31EE295 pKa = 4.08ITSFFNMNNRR305 pKa = 11.84TRR307 pKa = 11.84FPVYY311 pKa = 8.85WNDD314 pKa = 3.22YY315 pKa = 9.71RR316 pKa = 11.84SEE318 pKa = 4.1NKK320 pKa = 8.61DD321 pKa = 2.98TKK323 pKa = 10.57LTVDD327 pKa = 3.98EE328 pKa = 4.72DD329 pKa = 4.65TVSLSEE335 pKa = 4.19GADD338 pKa = 3.19LKK340 pKa = 11.24KK341 pKa = 10.78IMEE344 pKa = 4.32
MM1 pKa = 7.31EE2 pKa = 5.87AFRR5 pKa = 11.84MVSVTLRR12 pKa = 11.84EE13 pKa = 4.16TYY15 pKa = 10.57DD16 pKa = 3.5LSTKK20 pKa = 8.55QGKK23 pKa = 6.68MTLIGIHH30 pKa = 6.54IPTSNIIQRR39 pKa = 11.84LYY41 pKa = 10.48PGFCEE46 pKa = 3.99QYY48 pKa = 9.9KK49 pKa = 8.73MCRR52 pKa = 11.84INYY55 pKa = 7.55QNVRR59 pKa = 11.84IEE61 pKa = 4.26SASRR65 pKa = 11.84LPLDD69 pKa = 3.71PAGVGVNEE77 pKa = 4.54EE78 pKa = 4.42GLVSPEE84 pKa = 4.31DD85 pKa = 3.56VFNPILYY92 pKa = 9.92KK93 pKa = 10.64AVSNQSLSNLEE104 pKa = 3.55YY105 pKa = 10.48RR106 pKa = 11.84IRR108 pKa = 11.84GLRR111 pKa = 11.84DD112 pKa = 2.94QEE114 pKa = 4.25LSVDD118 pKa = 4.07GEE120 pKa = 4.47SATASNEE127 pKa = 3.75NVTFLDD133 pKa = 3.92DD134 pKa = 3.93EE135 pKa = 5.03FNVYY139 pKa = 10.57YY140 pKa = 10.7SILSNTRR147 pKa = 11.84GWKK150 pKa = 8.69VAHH153 pKa = 6.23PQAGLSMRR161 pKa = 11.84KK162 pKa = 9.14LRR164 pKa = 11.84PLVFEE169 pKa = 4.15KK170 pKa = 10.3WFSEE174 pKa = 4.24GVNGVNDD181 pKa = 3.18FTIPVDD187 pKa = 3.73SSHH190 pKa = 7.7LEE192 pKa = 3.91GEE194 pKa = 4.5TPVNYY199 pKa = 9.4IGQMPDD205 pKa = 2.29EE206 pKa = 4.6AMRR209 pKa = 11.84GRR211 pKa = 11.84PHH213 pKa = 7.84AMPAFNTKK221 pKa = 10.23YY222 pKa = 9.76LTGVQDD228 pKa = 3.5GSGAGVGKK236 pKa = 9.37FQQNGMGDD244 pKa = 4.18GLPGNAQVSMPILPTIYY261 pKa = 9.6TGLILVPPSRR271 pKa = 11.84NTTLYY276 pKa = 9.58YY277 pKa = 10.66RR278 pKa = 11.84MVVEE282 pKa = 4.47TVVSFWGVQTMDD294 pKa = 4.31EE295 pKa = 4.08ITSFFNMNNRR305 pKa = 11.84TRR307 pKa = 11.84FPVYY311 pKa = 8.85WNDD314 pKa = 3.22YY315 pKa = 9.71RR316 pKa = 11.84SEE318 pKa = 4.1NKK320 pKa = 8.61DD321 pKa = 2.98TKK323 pKa = 10.57LTVDD327 pKa = 3.98EE328 pKa = 4.72DD329 pKa = 4.65TVSLSEE335 pKa = 4.19GADD338 pKa = 3.19LKK340 pKa = 11.24KK341 pKa = 10.78IMEE344 pKa = 4.32
Molecular weight: 38.65 kDa
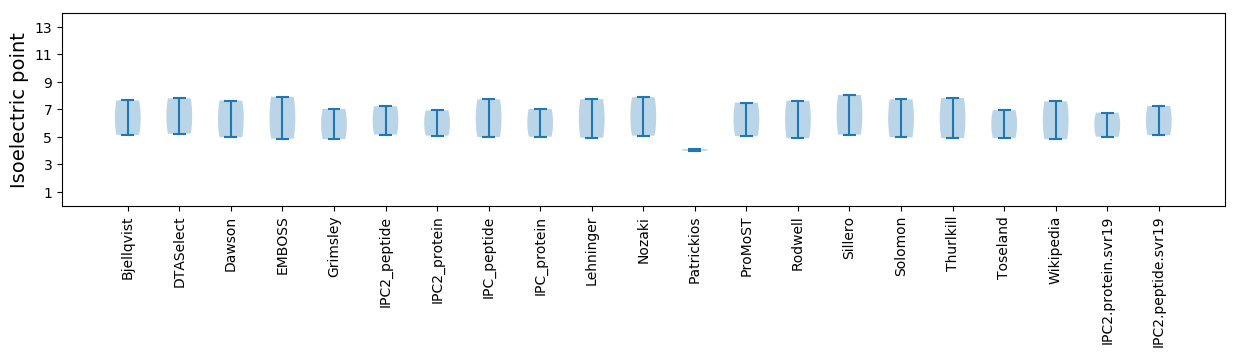
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A161IB94|A0A161IB94_9VIRU Putative capsid protein OS=Bovine faeces associated smacovirus 3 OX=1843751 PE=4 SV=1
MM1 pKa = 7.5GKK3 pKa = 10.37YY4 pKa = 10.17SVTISADD11 pKa = 2.82LWKK14 pKa = 10.66EE15 pKa = 3.49RR16 pKa = 11.84DD17 pKa = 3.72VVRR20 pKa = 11.84LLDD23 pKa = 3.62MVDD26 pKa = 3.03LRR28 pKa = 11.84EE29 pKa = 4.01YY30 pKa = 10.88YY31 pKa = 9.69IGRR34 pKa = 11.84EE35 pKa = 3.79IGKK38 pKa = 9.87GGYY41 pKa = 8.2HH42 pKa = 7.3HH43 pKa = 6.56YY44 pKa = 10.48QCAIDD49 pKa = 4.26CGGDD53 pKa = 3.3LEE55 pKa = 4.86RR56 pKa = 11.84FNGQHH61 pKa = 4.76QLGWHH66 pKa = 6.5IEE68 pKa = 4.26DD69 pKa = 3.85CASWDD74 pKa = 3.37KK75 pKa = 11.02LVRR78 pKa = 11.84YY79 pKa = 8.65CRR81 pKa = 11.84KK82 pKa = 10.24GGDD85 pKa = 3.21YY86 pKa = 10.36RR87 pKa = 11.84YY88 pKa = 9.89IGDD91 pKa = 4.1SIEE94 pKa = 4.09EE95 pKa = 3.49QSYY98 pKa = 10.28RR99 pKa = 11.84SRR101 pKa = 11.84GTTAVGHH108 pKa = 6.54TIDD111 pKa = 3.74THH113 pKa = 7.07LKK115 pKa = 8.72KK116 pKa = 10.89QNDD119 pKa = 4.0RR120 pKa = 11.84QISICVDD127 pKa = 3.27TKK129 pKa = 11.0GGSGKK134 pKa = 6.63TTHH137 pKa = 6.72GYY139 pKa = 10.11DD140 pKa = 3.11RR141 pKa = 11.84SRR143 pKa = 11.84TGEE146 pKa = 4.06YY147 pKa = 10.36FVVPRR152 pKa = 11.84NAEE155 pKa = 3.9TPTRR159 pKa = 11.84IMDD162 pKa = 3.9YY163 pKa = 11.27VAMNYY168 pKa = 10.42DD169 pKa = 3.94NEE171 pKa = 4.18PVIWIDD177 pKa = 3.56LPRR180 pKa = 11.84TRR182 pKa = 11.84KK183 pKa = 8.98IDD185 pKa = 3.4KK186 pKa = 10.46GLATILEE193 pKa = 4.18DD194 pKa = 3.59MKK196 pKa = 11.41DD197 pKa = 3.52GLIYY201 pKa = 9.39SAKK204 pKa = 9.78YY205 pKa = 8.67EE206 pKa = 4.17GQVRR210 pKa = 11.84HH211 pKa = 5.56IKK213 pKa = 9.54GVKK216 pKa = 9.55VLVTTNHH223 pKa = 6.23YY224 pKa = 9.91IEE226 pKa = 4.71KK227 pKa = 9.52PAYY230 pKa = 9.0KK231 pKa = 10.22LLSADD236 pKa = 3.23RR237 pKa = 11.84WDD239 pKa = 3.9VFTPPTKK246 pKa = 10.75AGDD249 pKa = 3.8SRR251 pKa = 11.84DD252 pKa = 3.77GNTGLSPLGGG262 pKa = 3.52
MM1 pKa = 7.5GKK3 pKa = 10.37YY4 pKa = 10.17SVTISADD11 pKa = 2.82LWKK14 pKa = 10.66EE15 pKa = 3.49RR16 pKa = 11.84DD17 pKa = 3.72VVRR20 pKa = 11.84LLDD23 pKa = 3.62MVDD26 pKa = 3.03LRR28 pKa = 11.84EE29 pKa = 4.01YY30 pKa = 10.88YY31 pKa = 9.69IGRR34 pKa = 11.84EE35 pKa = 3.79IGKK38 pKa = 9.87GGYY41 pKa = 8.2HH42 pKa = 7.3HH43 pKa = 6.56YY44 pKa = 10.48QCAIDD49 pKa = 4.26CGGDD53 pKa = 3.3LEE55 pKa = 4.86RR56 pKa = 11.84FNGQHH61 pKa = 4.76QLGWHH66 pKa = 6.5IEE68 pKa = 4.26DD69 pKa = 3.85CASWDD74 pKa = 3.37KK75 pKa = 11.02LVRR78 pKa = 11.84YY79 pKa = 8.65CRR81 pKa = 11.84KK82 pKa = 10.24GGDD85 pKa = 3.21YY86 pKa = 10.36RR87 pKa = 11.84YY88 pKa = 9.89IGDD91 pKa = 4.1SIEE94 pKa = 4.09EE95 pKa = 3.49QSYY98 pKa = 10.28RR99 pKa = 11.84SRR101 pKa = 11.84GTTAVGHH108 pKa = 6.54TIDD111 pKa = 3.74THH113 pKa = 7.07LKK115 pKa = 8.72KK116 pKa = 10.89QNDD119 pKa = 4.0RR120 pKa = 11.84QISICVDD127 pKa = 3.27TKK129 pKa = 11.0GGSGKK134 pKa = 6.63TTHH137 pKa = 6.72GYY139 pKa = 10.11DD140 pKa = 3.11RR141 pKa = 11.84SRR143 pKa = 11.84TGEE146 pKa = 4.06YY147 pKa = 10.36FVVPRR152 pKa = 11.84NAEE155 pKa = 3.9TPTRR159 pKa = 11.84IMDD162 pKa = 3.9YY163 pKa = 11.27VAMNYY168 pKa = 10.42DD169 pKa = 3.94NEE171 pKa = 4.18PVIWIDD177 pKa = 3.56LPRR180 pKa = 11.84TRR182 pKa = 11.84KK183 pKa = 8.98IDD185 pKa = 3.4KK186 pKa = 10.46GLATILEE193 pKa = 4.18DD194 pKa = 3.59MKK196 pKa = 11.41DD197 pKa = 3.52GLIYY201 pKa = 9.39SAKK204 pKa = 9.78YY205 pKa = 8.67EE206 pKa = 4.17GQVRR210 pKa = 11.84HH211 pKa = 5.56IKK213 pKa = 9.54GVKK216 pKa = 9.55VLVTTNHH223 pKa = 6.23YY224 pKa = 9.91IEE226 pKa = 4.71KK227 pKa = 9.52PAYY230 pKa = 9.0KK231 pKa = 10.22LLSADD236 pKa = 3.23RR237 pKa = 11.84WDD239 pKa = 3.9VFTPPTKK246 pKa = 10.75AGDD249 pKa = 3.8SRR251 pKa = 11.84DD252 pKa = 3.77GNTGLSPLGGG262 pKa = 3.52
Molecular weight: 29.79 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
606 |
262 |
344 |
303.0 |
34.22 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.125 ± 0.044 | 1.155 ± 0.453 |
6.931 ± 1.342 | 5.941 ± 0.589 |
2.805 ± 0.999 | 9.076 ± 0.97 |
2.145 ± 0.776 | 5.776 ± 0.659 |
5.116 ± 1.056 | 7.261 ± 0.465 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.135 ± 0.738 | 4.95 ± 1.372 |
4.455 ± 0.844 | 3.3 ± 0.378 |
6.436 ± 0.721 | 6.436 ± 0.887 |
6.766 ± 0.063 | 7.426 ± 0.794 |
1.485 ± 0.255 | 5.281 ± 0.727 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
