
Caulobacter sp. 17J65-9
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Caulobacterales; Caulobacteraceae; Caulobacter; unclassified Caulobacter
Average proteome isoelectric point is 6.59
Get precalculated fractions of proteins
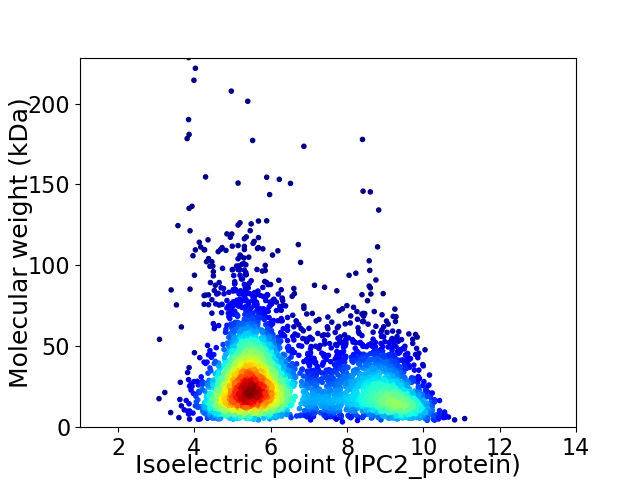
Virtual 2D-PAGE plot for 4292 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6B3UKC1|A0A6B3UKC1_9CAUL Uncharacterized protein (Fragment) OS=Caulobacter sp. 17J65-9 OX=2709382 GN=G3573_14915 PE=4 SV=1
MM1 pKa = 7.29SSRR4 pKa = 11.84RR5 pKa = 11.84GVLLLGSALGVSIAVLAAAVPNRR28 pKa = 11.84ALAADD33 pKa = 3.59EE34 pKa = 4.88CGAVPAGATPTVTCTVADD52 pKa = 3.7SPYY55 pKa = 10.6PGVQYY60 pKa = 11.08DD61 pKa = 3.9GVEE64 pKa = 4.16DD65 pKa = 3.88LTVVIQPDD73 pKa = 3.73VTVEE77 pKa = 3.75GLVRR81 pKa = 11.84VTGTGDD87 pKa = 3.54LAVNAQAGSSIDD99 pKa = 3.6NPTGAGIQVTSSGTGSVTAASVSAGYY125 pKa = 10.88GDD127 pKa = 4.07GVSVSGEE134 pKa = 3.52AGATVNVGTVSGDD147 pKa = 3.12WTGVAASSNLGSVTVTSTSVTAVEE171 pKa = 4.37GRR173 pKa = 11.84GIFAWARR180 pKa = 11.84AEE182 pKa = 4.09GEE184 pKa = 4.51TINITSGTVEE194 pKa = 4.38SYY196 pKa = 11.51DD197 pKa = 3.5EE198 pKa = 4.94GIYY201 pKa = 10.29AYY203 pKa = 10.42SGEE206 pKa = 4.35GGSADD211 pKa = 3.48VVVDD215 pKa = 3.71SATVTTTRR223 pKa = 11.84SSWATGIVAAASHH236 pKa = 6.8GDD238 pKa = 3.42VTVTSGSVTTAGDD251 pKa = 3.61YY252 pKa = 11.29SEE254 pKa = 5.82GILVKK259 pKa = 10.3AAEE262 pKa = 4.48GEE264 pKa = 4.62VYY266 pKa = 10.35VQSDD270 pKa = 4.15TVHH273 pKa = 5.97TAGDD277 pKa = 3.88YY278 pKa = 8.64STGIKK283 pKa = 10.28VEE285 pKa = 4.32GGLGPVEE292 pKa = 4.07LHH294 pKa = 6.56SGSITTDD301 pKa = 3.13GASASGIDD309 pKa = 3.64VYY311 pKa = 8.93TTSGAVTVTSDD322 pKa = 4.07EE323 pKa = 3.93IALNGGRR330 pKa = 11.84AVEE333 pKa = 4.31TTYY336 pKa = 11.49GEE338 pKa = 4.26YY339 pKa = 10.96GSYY342 pKa = 7.09TTYY345 pKa = 10.62GRR347 pKa = 11.84RR348 pKa = 11.84PVGIDD353 pKa = 3.09VRR355 pKa = 11.84STDD358 pKa = 3.34GAVTITSGDD367 pKa = 3.17ISVTDD372 pKa = 4.66GIGISVEE379 pKa = 3.99TAGNISITSTGTLLVADD396 pKa = 4.63GEE398 pKa = 4.79RR399 pKa = 11.84NGGYY403 pKa = 10.01GYY405 pKa = 11.05SSGIYY410 pKa = 9.82FDD412 pKa = 4.18SQNSGEE418 pKa = 4.4GEE420 pKa = 3.91PGGTVQVVNEE430 pKa = 4.0GRR432 pKa = 11.84IEE434 pKa = 3.74IHH436 pKa = 5.75GSNRR440 pKa = 11.84NGIYY444 pKa = 10.4GYY446 pKa = 10.95GSGSAVSITNAGEE459 pKa = 3.98IVGDD463 pKa = 3.81RR464 pKa = 11.84DD465 pKa = 3.28TTAIRR470 pKa = 11.84VHH472 pKa = 6.93NEE474 pKa = 3.42DD475 pKa = 3.58GGALSVTNQAGASIHH490 pKa = 6.96LYY492 pKa = 9.02DD493 pKa = 3.97TYY495 pKa = 11.13STGIHH500 pKa = 6.68AGTDD504 pKa = 3.22EE505 pKa = 4.51GALTITNAGSIVTEE519 pKa = 4.13GDD521 pKa = 3.3YY522 pKa = 11.35SSAITFHH529 pKa = 7.52SEE531 pKa = 3.46AGTVTLNSTGTIEE544 pKa = 4.02TSGYY548 pKa = 10.36GSGAIYY554 pKa = 9.8GHH556 pKa = 6.45SYY558 pKa = 11.08DD559 pKa = 4.15GDD561 pKa = 3.91VLVDD565 pKa = 3.45VASVHH570 pKa = 6.07VGGGSASGIDD580 pKa = 3.48VWSEE584 pKa = 3.92SGSVRR589 pKa = 11.84VNADD593 pKa = 3.37LVVTGSSCSEE603 pKa = 4.04GEE605 pKa = 4.38GCSGEE610 pKa = 4.1GDD612 pKa = 3.92SVGVSASSGTGNVTIDD628 pKa = 3.49VGSVTTGDD636 pKa = 3.9DD637 pKa = 3.61YY638 pKa = 12.01AVGVSAGMYY647 pKa = 10.43GGFLGDD653 pKa = 3.63AYY655 pKa = 10.79SGEE658 pKa = 4.21GEE660 pKa = 4.59YY661 pKa = 11.09VFGDD665 pKa = 3.39IQVTADD671 pKa = 3.29RR672 pKa = 11.84VTTSGDD678 pKa = 2.81WAAGIQVISGRR689 pKa = 11.84DD690 pKa = 3.6EE691 pKa = 4.6YY692 pKa = 10.94ISGPFGPPPIVRR704 pKa = 11.84TGAVDD709 pKa = 3.59IDD711 pKa = 3.6AGTIVTHH718 pKa = 6.6GSGSAGISVAAGSAEE733 pKa = 3.94DD734 pKa = 4.85VLGFWSGPMLGGEE747 pKa = 4.13GGVNPFALVMGPHH760 pKa = 7.1FGTTGDD766 pKa = 3.19VDD768 pKa = 3.75VDD770 pKa = 4.21VVSVSTDD777 pKa = 2.52GDD779 pKa = 4.04GAVGVYY785 pKa = 10.16ASSGFTLGFYY795 pKa = 11.29GEE797 pKa = 4.53GEE799 pKa = 4.26YY800 pKa = 11.14GGAEE804 pKa = 3.95PGGDD808 pKa = 3.48VIVTSGAIEE817 pKa = 4.0TAGDD821 pKa = 3.64GAIGIVARR829 pKa = 11.84GVAGGVTIDD838 pKa = 3.53SGTIEE843 pKa = 4.11TSGDD847 pKa = 3.58YY848 pKa = 11.05ASGIVANNGGVVFTRR863 pKa = 11.84VEE865 pKa = 4.09IEE867 pKa = 3.88PEE869 pKa = 4.24FYY871 pKa = 8.38TTVPSFVSGGLIDD884 pKa = 3.74VTSDD888 pKa = 3.17KK889 pKa = 10.61IYY891 pKa = 10.73TSGEE895 pKa = 3.81SSAGITAVGMGGGVTIDD912 pKa = 3.66SGIIDD917 pKa = 3.44TWGAYY922 pKa = 10.33ANGIQVMAIGGDD934 pKa = 3.46VSVDD938 pKa = 3.15STRR941 pKa = 11.84IIARR945 pKa = 11.84GDD947 pKa = 3.13RR948 pKa = 11.84SGGILINTANLDD960 pKa = 3.72SFFGEE965 pKa = 4.52GEE967 pKa = 4.03YY968 pKa = 11.08GATFVAGDD976 pKa = 3.19IHH978 pKa = 5.87VTSGTIRR985 pKa = 11.84TEE987 pKa = 3.98GGSADD992 pKa = 4.24GIAVFSGYY1000 pKa = 11.25AFFGEE1005 pKa = 4.63GEE1007 pKa = 4.26YY1008 pKa = 11.21GGGDD1012 pKa = 3.34VDD1014 pKa = 3.56ITSGNITTLGDD1025 pKa = 3.44DD1026 pKa = 3.5SAGIRR1031 pKa = 11.84VTAYY1035 pKa = 10.22GRR1037 pKa = 11.84YY1038 pKa = 7.93TYY1040 pKa = 11.23GDD1042 pKa = 3.62DD1043 pKa = 3.87EE1044 pKa = 4.38SWEE1047 pKa = 3.98IEE1049 pKa = 3.96GDD1051 pKa = 3.89VTVHH1055 pKa = 6.04SGAVITSGDD1064 pKa = 3.74YY1065 pKa = 10.56SDD1067 pKa = 5.59GIVVEE1072 pKa = 4.28ATLGDD1077 pKa = 3.91VNVTANATATSGDD1090 pKa = 3.51SSRR1093 pKa = 11.84GIDD1096 pKa = 3.05IDD1098 pKa = 4.02ADD1100 pKa = 3.68NGDD1103 pKa = 3.76VVVNATAVTTTGGRR1117 pKa = 11.84KK1118 pKa = 9.37ANEE1121 pKa = 3.42VSYY1124 pKa = 10.67YY1125 pKa = 10.95DD1126 pKa = 4.06SYY1128 pKa = 10.51TEE1130 pKa = 3.51SWVYY1134 pKa = 7.95VTEE1137 pKa = 4.2YY1138 pKa = 10.92GHH1140 pKa = 7.07ASDD1143 pKa = 5.49GIRR1146 pKa = 11.84VEE1148 pKa = 4.24TEE1150 pKa = 3.79YY1151 pKa = 11.78GDD1153 pKa = 3.7VTINAQNVTVSGDD1166 pKa = 3.12QATGVLVDD1174 pKa = 4.12SEE1176 pKa = 4.52YY1177 pKa = 11.66GDD1179 pKa = 3.41VKK1181 pKa = 11.25LNLGSVRR1188 pKa = 11.84AQGEE1192 pKa = 3.99AGDD1195 pKa = 4.14AVHH1198 pKa = 6.71VYY1200 pKa = 10.19AYY1202 pKa = 10.14EE1203 pKa = 4.09GDD1205 pKa = 3.23IGVNISGQVTSANGAGVHH1223 pKa = 6.39AYY1225 pKa = 9.8SYY1227 pKa = 11.15YY1228 pKa = 10.93GGVDD1232 pKa = 3.27VKK1234 pKa = 10.68LAAGSRR1240 pKa = 11.84VQGGTVGIYY1249 pKa = 9.98AYY1251 pKa = 10.48SDD1253 pKa = 3.11TGTTITINGTVSAASGLAIDD1273 pKa = 4.46VKK1275 pKa = 11.19GLGAATIHH1283 pKa = 6.03NLSNTVIGGVSLTDD1297 pKa = 3.76NDD1299 pKa = 5.06DD1300 pKa = 4.28LFDD1303 pKa = 3.75NTGTFQATIDD1313 pKa = 3.77SQFGEE1318 pKa = 4.62GVDD1321 pKa = 4.59LFDD1324 pKa = 4.51NTGLLQILPGSSTPGTIVFQGLEE1347 pKa = 4.1TFDD1350 pKa = 3.55NAGVVDD1356 pKa = 5.02LANGHH1361 pKa = 6.61AGDD1364 pKa = 4.29VFDD1367 pKa = 6.09LSSATWNGEE1376 pKa = 3.82AGSTLEE1382 pKa = 4.67LDD1384 pKa = 3.55ISGSGSDD1391 pKa = 3.53TLLVDD1396 pKa = 4.44DD1397 pKa = 4.9ATGHH1401 pKa = 4.52TTIVLSGAGGGIGQALEE1418 pKa = 4.37LVVSNSDD1425 pKa = 3.15EE1426 pKa = 4.25TGNEE1430 pKa = 3.69FTLDD1434 pKa = 3.34GDD1436 pKa = 3.78TGFVKK1441 pKa = 10.81FIMDD1445 pKa = 3.87FDD1447 pKa = 4.51AATDD1451 pKa = 3.21TWSAVGTADD1460 pKa = 3.69TEE1462 pKa = 4.33VFEE1465 pKa = 5.03PGRR1468 pKa = 11.84FLTGAQNFWNEE1479 pKa = 4.02TGDD1482 pKa = 3.2AWSARR1487 pKa = 11.84MTEE1490 pKa = 4.01LRR1492 pKa = 11.84DD1493 pKa = 3.26IGSDD1497 pKa = 3.09RR1498 pKa = 11.84GQGAQGWVQAFGGEE1512 pKa = 4.06QSEE1515 pKa = 4.46DD1516 pKa = 3.07RR1517 pKa = 11.84GTEE1520 pKa = 3.64RR1521 pKa = 11.84FTVGGAPSDD1530 pKa = 3.67VNLAYY1535 pKa = 9.43EE1536 pKa = 3.7QSYY1539 pKa = 10.84RR1540 pKa = 11.84GLQTGVDD1547 pKa = 3.72WSTGDD1552 pKa = 3.27TVLGVTIGFGDD1563 pKa = 3.65SEE1565 pKa = 4.25QRR1567 pKa = 11.84MASGNSAEE1575 pKa = 4.0FGGFNFGAYY1584 pKa = 9.89GGWNPGSFFVNGLVKK1599 pKa = 10.54ADD1601 pKa = 3.59VYY1603 pKa = 10.02KK1604 pKa = 10.68TRR1606 pKa = 11.84LGLRR1610 pKa = 11.84TLQASEE1616 pKa = 4.25EE1617 pKa = 4.24VDD1619 pKa = 3.16GRR1621 pKa = 11.84TWGAKK1626 pKa = 9.9AEE1628 pKa = 4.32AGWRR1632 pKa = 11.84VGGDD1636 pKa = 2.75GWFFEE1641 pKa = 4.61PSARR1645 pKa = 11.84LSWTDD1650 pKa = 2.53VDD1652 pKa = 3.68MDD1654 pKa = 3.98GFSAAGGTVEE1664 pKa = 5.76FGDD1667 pKa = 3.82ASSLLGEE1674 pKa = 4.25AGARR1678 pKa = 11.84FGTHH1682 pKa = 4.32WTNGVVDD1689 pKa = 4.81LVPYY1693 pKa = 10.6LGLFAVDD1700 pKa = 4.49EE1701 pKa = 4.59FDD1703 pKa = 5.96GEE1705 pKa = 4.27NQTAFSIAGDD1715 pKa = 3.79TFRR1718 pKa = 11.84LTDD1721 pKa = 3.61EE1722 pKa = 4.25AAGAHH1727 pKa = 5.55GRR1729 pKa = 11.84VDD1731 pKa = 4.27FGVTAVTFHH1740 pKa = 6.28GLEE1743 pKa = 4.16GFVKK1747 pKa = 10.91GEE1749 pKa = 3.93ALVGGDD1755 pKa = 3.53AEE1757 pKa = 5.05GYY1759 pKa = 8.32SARR1762 pKa = 11.84LGVRR1766 pKa = 11.84WRR1768 pKa = 11.84WW1769 pKa = 3.03
MM1 pKa = 7.29SSRR4 pKa = 11.84RR5 pKa = 11.84GVLLLGSALGVSIAVLAAAVPNRR28 pKa = 11.84ALAADD33 pKa = 3.59EE34 pKa = 4.88CGAVPAGATPTVTCTVADD52 pKa = 3.7SPYY55 pKa = 10.6PGVQYY60 pKa = 11.08DD61 pKa = 3.9GVEE64 pKa = 4.16DD65 pKa = 3.88LTVVIQPDD73 pKa = 3.73VTVEE77 pKa = 3.75GLVRR81 pKa = 11.84VTGTGDD87 pKa = 3.54LAVNAQAGSSIDD99 pKa = 3.6NPTGAGIQVTSSGTGSVTAASVSAGYY125 pKa = 10.88GDD127 pKa = 4.07GVSVSGEE134 pKa = 3.52AGATVNVGTVSGDD147 pKa = 3.12WTGVAASSNLGSVTVTSTSVTAVEE171 pKa = 4.37GRR173 pKa = 11.84GIFAWARR180 pKa = 11.84AEE182 pKa = 4.09GEE184 pKa = 4.51TINITSGTVEE194 pKa = 4.38SYY196 pKa = 11.51DD197 pKa = 3.5EE198 pKa = 4.94GIYY201 pKa = 10.29AYY203 pKa = 10.42SGEE206 pKa = 4.35GGSADD211 pKa = 3.48VVVDD215 pKa = 3.71SATVTTTRR223 pKa = 11.84SSWATGIVAAASHH236 pKa = 6.8GDD238 pKa = 3.42VTVTSGSVTTAGDD251 pKa = 3.61YY252 pKa = 11.29SEE254 pKa = 5.82GILVKK259 pKa = 10.3AAEE262 pKa = 4.48GEE264 pKa = 4.62VYY266 pKa = 10.35VQSDD270 pKa = 4.15TVHH273 pKa = 5.97TAGDD277 pKa = 3.88YY278 pKa = 8.64STGIKK283 pKa = 10.28VEE285 pKa = 4.32GGLGPVEE292 pKa = 4.07LHH294 pKa = 6.56SGSITTDD301 pKa = 3.13GASASGIDD309 pKa = 3.64VYY311 pKa = 8.93TTSGAVTVTSDD322 pKa = 4.07EE323 pKa = 3.93IALNGGRR330 pKa = 11.84AVEE333 pKa = 4.31TTYY336 pKa = 11.49GEE338 pKa = 4.26YY339 pKa = 10.96GSYY342 pKa = 7.09TTYY345 pKa = 10.62GRR347 pKa = 11.84RR348 pKa = 11.84PVGIDD353 pKa = 3.09VRR355 pKa = 11.84STDD358 pKa = 3.34GAVTITSGDD367 pKa = 3.17ISVTDD372 pKa = 4.66GIGISVEE379 pKa = 3.99TAGNISITSTGTLLVADD396 pKa = 4.63GEE398 pKa = 4.79RR399 pKa = 11.84NGGYY403 pKa = 10.01GYY405 pKa = 11.05SSGIYY410 pKa = 9.82FDD412 pKa = 4.18SQNSGEE418 pKa = 4.4GEE420 pKa = 3.91PGGTVQVVNEE430 pKa = 4.0GRR432 pKa = 11.84IEE434 pKa = 3.74IHH436 pKa = 5.75GSNRR440 pKa = 11.84NGIYY444 pKa = 10.4GYY446 pKa = 10.95GSGSAVSITNAGEE459 pKa = 3.98IVGDD463 pKa = 3.81RR464 pKa = 11.84DD465 pKa = 3.28TTAIRR470 pKa = 11.84VHH472 pKa = 6.93NEE474 pKa = 3.42DD475 pKa = 3.58GGALSVTNQAGASIHH490 pKa = 6.96LYY492 pKa = 9.02DD493 pKa = 3.97TYY495 pKa = 11.13STGIHH500 pKa = 6.68AGTDD504 pKa = 3.22EE505 pKa = 4.51GALTITNAGSIVTEE519 pKa = 4.13GDD521 pKa = 3.3YY522 pKa = 11.35SSAITFHH529 pKa = 7.52SEE531 pKa = 3.46AGTVTLNSTGTIEE544 pKa = 4.02TSGYY548 pKa = 10.36GSGAIYY554 pKa = 9.8GHH556 pKa = 6.45SYY558 pKa = 11.08DD559 pKa = 4.15GDD561 pKa = 3.91VLVDD565 pKa = 3.45VASVHH570 pKa = 6.07VGGGSASGIDD580 pKa = 3.48VWSEE584 pKa = 3.92SGSVRR589 pKa = 11.84VNADD593 pKa = 3.37LVVTGSSCSEE603 pKa = 4.04GEE605 pKa = 4.38GCSGEE610 pKa = 4.1GDD612 pKa = 3.92SVGVSASSGTGNVTIDD628 pKa = 3.49VGSVTTGDD636 pKa = 3.9DD637 pKa = 3.61YY638 pKa = 12.01AVGVSAGMYY647 pKa = 10.43GGFLGDD653 pKa = 3.63AYY655 pKa = 10.79SGEE658 pKa = 4.21GEE660 pKa = 4.59YY661 pKa = 11.09VFGDD665 pKa = 3.39IQVTADD671 pKa = 3.29RR672 pKa = 11.84VTTSGDD678 pKa = 2.81WAAGIQVISGRR689 pKa = 11.84DD690 pKa = 3.6EE691 pKa = 4.6YY692 pKa = 10.94ISGPFGPPPIVRR704 pKa = 11.84TGAVDD709 pKa = 3.59IDD711 pKa = 3.6AGTIVTHH718 pKa = 6.6GSGSAGISVAAGSAEE733 pKa = 3.94DD734 pKa = 4.85VLGFWSGPMLGGEE747 pKa = 4.13GGVNPFALVMGPHH760 pKa = 7.1FGTTGDD766 pKa = 3.19VDD768 pKa = 3.75VDD770 pKa = 4.21VVSVSTDD777 pKa = 2.52GDD779 pKa = 4.04GAVGVYY785 pKa = 10.16ASSGFTLGFYY795 pKa = 11.29GEE797 pKa = 4.53GEE799 pKa = 4.26YY800 pKa = 11.14GGAEE804 pKa = 3.95PGGDD808 pKa = 3.48VIVTSGAIEE817 pKa = 4.0TAGDD821 pKa = 3.64GAIGIVARR829 pKa = 11.84GVAGGVTIDD838 pKa = 3.53SGTIEE843 pKa = 4.11TSGDD847 pKa = 3.58YY848 pKa = 11.05ASGIVANNGGVVFTRR863 pKa = 11.84VEE865 pKa = 4.09IEE867 pKa = 3.88PEE869 pKa = 4.24FYY871 pKa = 8.38TTVPSFVSGGLIDD884 pKa = 3.74VTSDD888 pKa = 3.17KK889 pKa = 10.61IYY891 pKa = 10.73TSGEE895 pKa = 3.81SSAGITAVGMGGGVTIDD912 pKa = 3.66SGIIDD917 pKa = 3.44TWGAYY922 pKa = 10.33ANGIQVMAIGGDD934 pKa = 3.46VSVDD938 pKa = 3.15STRR941 pKa = 11.84IIARR945 pKa = 11.84GDD947 pKa = 3.13RR948 pKa = 11.84SGGILINTANLDD960 pKa = 3.72SFFGEE965 pKa = 4.52GEE967 pKa = 4.03YY968 pKa = 11.08GATFVAGDD976 pKa = 3.19IHH978 pKa = 5.87VTSGTIRR985 pKa = 11.84TEE987 pKa = 3.98GGSADD992 pKa = 4.24GIAVFSGYY1000 pKa = 11.25AFFGEE1005 pKa = 4.63GEE1007 pKa = 4.26YY1008 pKa = 11.21GGGDD1012 pKa = 3.34VDD1014 pKa = 3.56ITSGNITTLGDD1025 pKa = 3.44DD1026 pKa = 3.5SAGIRR1031 pKa = 11.84VTAYY1035 pKa = 10.22GRR1037 pKa = 11.84YY1038 pKa = 7.93TYY1040 pKa = 11.23GDD1042 pKa = 3.62DD1043 pKa = 3.87EE1044 pKa = 4.38SWEE1047 pKa = 3.98IEE1049 pKa = 3.96GDD1051 pKa = 3.89VTVHH1055 pKa = 6.04SGAVITSGDD1064 pKa = 3.74YY1065 pKa = 10.56SDD1067 pKa = 5.59GIVVEE1072 pKa = 4.28ATLGDD1077 pKa = 3.91VNVTANATATSGDD1090 pKa = 3.51SSRR1093 pKa = 11.84GIDD1096 pKa = 3.05IDD1098 pKa = 4.02ADD1100 pKa = 3.68NGDD1103 pKa = 3.76VVVNATAVTTTGGRR1117 pKa = 11.84KK1118 pKa = 9.37ANEE1121 pKa = 3.42VSYY1124 pKa = 10.67YY1125 pKa = 10.95DD1126 pKa = 4.06SYY1128 pKa = 10.51TEE1130 pKa = 3.51SWVYY1134 pKa = 7.95VTEE1137 pKa = 4.2YY1138 pKa = 10.92GHH1140 pKa = 7.07ASDD1143 pKa = 5.49GIRR1146 pKa = 11.84VEE1148 pKa = 4.24TEE1150 pKa = 3.79YY1151 pKa = 11.78GDD1153 pKa = 3.7VTINAQNVTVSGDD1166 pKa = 3.12QATGVLVDD1174 pKa = 4.12SEE1176 pKa = 4.52YY1177 pKa = 11.66GDD1179 pKa = 3.41VKK1181 pKa = 11.25LNLGSVRR1188 pKa = 11.84AQGEE1192 pKa = 3.99AGDD1195 pKa = 4.14AVHH1198 pKa = 6.71VYY1200 pKa = 10.19AYY1202 pKa = 10.14EE1203 pKa = 4.09GDD1205 pKa = 3.23IGVNISGQVTSANGAGVHH1223 pKa = 6.39AYY1225 pKa = 9.8SYY1227 pKa = 11.15YY1228 pKa = 10.93GGVDD1232 pKa = 3.27VKK1234 pKa = 10.68LAAGSRR1240 pKa = 11.84VQGGTVGIYY1249 pKa = 9.98AYY1251 pKa = 10.48SDD1253 pKa = 3.11TGTTITINGTVSAASGLAIDD1273 pKa = 4.46VKK1275 pKa = 11.19GLGAATIHH1283 pKa = 6.03NLSNTVIGGVSLTDD1297 pKa = 3.76NDD1299 pKa = 5.06DD1300 pKa = 4.28LFDD1303 pKa = 3.75NTGTFQATIDD1313 pKa = 3.77SQFGEE1318 pKa = 4.62GVDD1321 pKa = 4.59LFDD1324 pKa = 4.51NTGLLQILPGSSTPGTIVFQGLEE1347 pKa = 4.1TFDD1350 pKa = 3.55NAGVVDD1356 pKa = 5.02LANGHH1361 pKa = 6.61AGDD1364 pKa = 4.29VFDD1367 pKa = 6.09LSSATWNGEE1376 pKa = 3.82AGSTLEE1382 pKa = 4.67LDD1384 pKa = 3.55ISGSGSDD1391 pKa = 3.53TLLVDD1396 pKa = 4.44DD1397 pKa = 4.9ATGHH1401 pKa = 4.52TTIVLSGAGGGIGQALEE1418 pKa = 4.37LVVSNSDD1425 pKa = 3.15EE1426 pKa = 4.25TGNEE1430 pKa = 3.69FTLDD1434 pKa = 3.34GDD1436 pKa = 3.78TGFVKK1441 pKa = 10.81FIMDD1445 pKa = 3.87FDD1447 pKa = 4.51AATDD1451 pKa = 3.21TWSAVGTADD1460 pKa = 3.69TEE1462 pKa = 4.33VFEE1465 pKa = 5.03PGRR1468 pKa = 11.84FLTGAQNFWNEE1479 pKa = 4.02TGDD1482 pKa = 3.2AWSARR1487 pKa = 11.84MTEE1490 pKa = 4.01LRR1492 pKa = 11.84DD1493 pKa = 3.26IGSDD1497 pKa = 3.09RR1498 pKa = 11.84GQGAQGWVQAFGGEE1512 pKa = 4.06QSEE1515 pKa = 4.46DD1516 pKa = 3.07RR1517 pKa = 11.84GTEE1520 pKa = 3.64RR1521 pKa = 11.84FTVGGAPSDD1530 pKa = 3.67VNLAYY1535 pKa = 9.43EE1536 pKa = 3.7QSYY1539 pKa = 10.84RR1540 pKa = 11.84GLQTGVDD1547 pKa = 3.72WSTGDD1552 pKa = 3.27TVLGVTIGFGDD1563 pKa = 3.65SEE1565 pKa = 4.25QRR1567 pKa = 11.84MASGNSAEE1575 pKa = 4.0FGGFNFGAYY1584 pKa = 9.89GGWNPGSFFVNGLVKK1599 pKa = 10.54ADD1601 pKa = 3.59VYY1603 pKa = 10.02KK1604 pKa = 10.68TRR1606 pKa = 11.84LGLRR1610 pKa = 11.84TLQASEE1616 pKa = 4.25EE1617 pKa = 4.24VDD1619 pKa = 3.16GRR1621 pKa = 11.84TWGAKK1626 pKa = 9.9AEE1628 pKa = 4.32AGWRR1632 pKa = 11.84VGGDD1636 pKa = 2.75GWFFEE1641 pKa = 4.61PSARR1645 pKa = 11.84LSWTDD1650 pKa = 2.53VDD1652 pKa = 3.68MDD1654 pKa = 3.98GFSAAGGTVEE1664 pKa = 5.76FGDD1667 pKa = 3.82ASSLLGEE1674 pKa = 4.25AGARR1678 pKa = 11.84FGTHH1682 pKa = 4.32WTNGVVDD1689 pKa = 4.81LVPYY1693 pKa = 10.6LGLFAVDD1700 pKa = 4.49EE1701 pKa = 4.59FDD1703 pKa = 5.96GEE1705 pKa = 4.27NQTAFSIAGDD1715 pKa = 3.79TFRR1718 pKa = 11.84LTDD1721 pKa = 3.61EE1722 pKa = 4.25AAGAHH1727 pKa = 5.55GRR1729 pKa = 11.84VDD1731 pKa = 4.27FGVTAVTFHH1740 pKa = 6.28GLEE1743 pKa = 4.16GFVKK1747 pKa = 10.91GEE1749 pKa = 3.93ALVGGDD1755 pKa = 3.53AEE1757 pKa = 5.05GYY1759 pKa = 8.32SARR1762 pKa = 11.84LGVRR1766 pKa = 11.84WRR1768 pKa = 11.84WW1769 pKa = 3.03
Molecular weight: 178.34 kDa
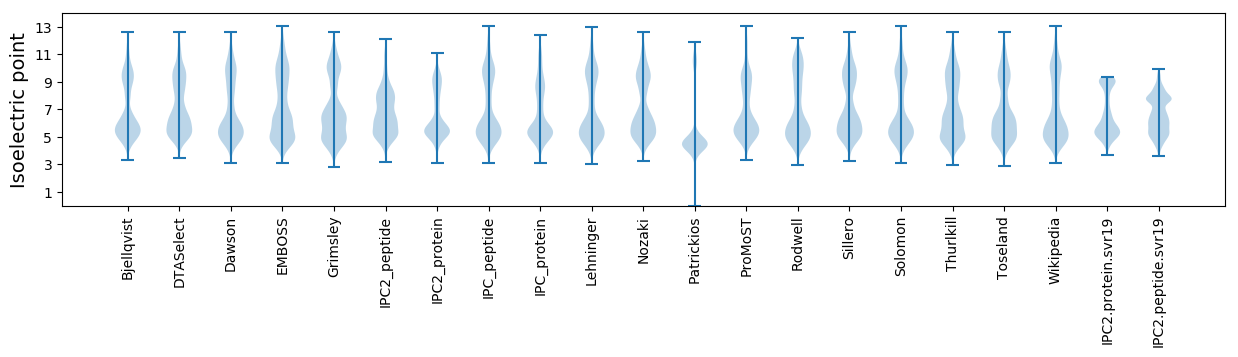
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6B3USG0|A0A6B3USG0_9CAUL Alpha/beta hydrolase OS=Caulobacter sp. 17J65-9 OX=2709382 GN=G3573_16210 PE=4 SV=1
AA1 pKa = 7.78SSAQSRR7 pKa = 11.84SAQARR12 pKa = 11.84GTRR15 pKa = 11.84RR16 pKa = 11.84WPPVIALSAAAAVSAGLWVAVVAAVRR42 pKa = 11.84ALLL45 pKa = 3.85
AA1 pKa = 7.78SSAQSRR7 pKa = 11.84SAQARR12 pKa = 11.84GTRR15 pKa = 11.84RR16 pKa = 11.84WPPVIALSAAAAVSAGLWVAVVAAVRR42 pKa = 11.84ALLL45 pKa = 3.85
Molecular weight: 4.52 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1247758 |
35 |
2282 |
290.7 |
31.25 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.464 ± 0.058 | 0.745 ± 0.014 |
5.869 ± 0.035 | 5.817 ± 0.04 |
3.554 ± 0.022 | 9.155 ± 0.051 |
1.737 ± 0.017 | 3.584 ± 0.022 |
3.063 ± 0.034 | 9.963 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.091 ± 0.017 | 2.134 ± 0.024 |
5.529 ± 0.032 | 2.975 ± 0.022 |
7.591 ± 0.039 | 4.786 ± 0.03 |
5.123 ± 0.042 | 8.106 ± 0.032 |
1.566 ± 0.017 | 2.146 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
