
Mobiluncus mulieris ATCC 35239
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Actinomycetales; Actinomycetaceae; Mobiluncus; Mobiluncus mulieris
Average proteome isoelectric point is 6.61
Get precalculated fractions of proteins
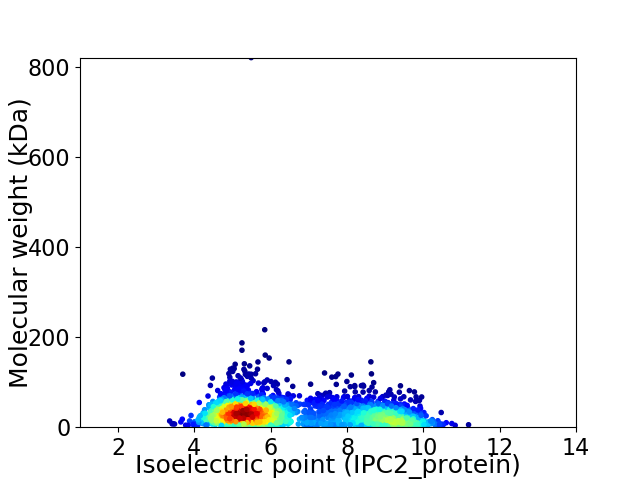
Virtual 2D-PAGE plot for 2379 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
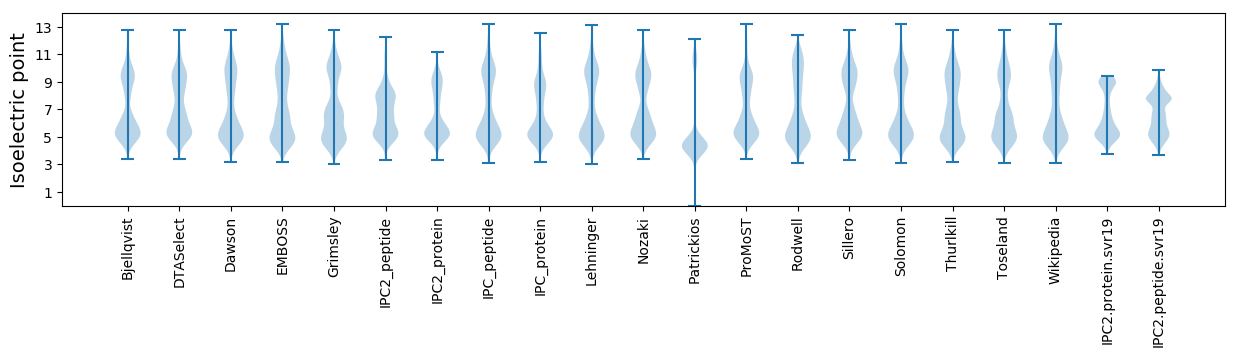
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E0QPV6|E0QPV6_9ACTO DEAD/DEAH box helicase OS=Mobiluncus mulieris ATCC 35239 OX=871571 GN=HMPREF0580_0883 PE=4 SV=1
MM1 pKa = 8.04SEE3 pKa = 4.45EE4 pKa = 4.1YY5 pKa = 10.16TPQLLTSEE13 pKa = 5.17DD14 pKa = 3.94VFTVQFPATKK24 pKa = 10.14FRR26 pKa = 11.84DD27 pKa = 4.43GYY29 pKa = 10.67DD30 pKa = 3.03QNQIDD35 pKa = 4.67DD36 pKa = 4.11YY37 pKa = 10.39LDD39 pKa = 3.29EE40 pKa = 4.38VVRR43 pKa = 11.84VLSYY47 pKa = 11.4YY48 pKa = 9.85EE49 pKa = 4.31ALSAAPEE56 pKa = 4.22AEE58 pKa = 3.8VDD60 pKa = 3.41LTYY63 pKa = 9.95ITVRR67 pKa = 11.84GVDD70 pKa = 3.47VRR72 pKa = 11.84EE73 pKa = 3.62VDD75 pKa = 3.92FDD77 pKa = 3.77YY78 pKa = 11.3TRR80 pKa = 11.84MRR82 pKa = 11.84VGYY85 pKa = 9.8DD86 pKa = 2.92QDD88 pKa = 4.8AVDD91 pKa = 5.52DD92 pKa = 4.27YY93 pKa = 11.24LDD95 pKa = 3.46QVAATLEE102 pKa = 4.35AYY104 pKa = 9.08EE105 pKa = 4.16NAYY108 pKa = 9.96GIPPSDD114 pKa = 3.18QRR116 pKa = 11.84YY117 pKa = 8.28QVLQVDD123 pKa = 5.36GPALEE128 pKa = 5.45AEE130 pKa = 4.7AAQQAAAAGEE140 pKa = 4.2LPAEE144 pKa = 4.37VSAPVALGDD153 pKa = 4.09SAAGGAPVTAGNMLSLGASVGGAVPLDD180 pKa = 4.1AAPSAPDD187 pKa = 3.19KK188 pKa = 10.87AASLGASEE196 pKa = 4.63TAPNSPGYY204 pKa = 10.39SSVLGTPPARR214 pKa = 11.84QPGVGEE220 pKa = 4.17FNEE223 pKa = 4.33APQYY227 pKa = 10.84PGDD230 pKa = 4.0SLDD233 pKa = 3.15WMQAPGQSPASSPQNYY249 pKa = 8.69PDD251 pKa = 4.1YY252 pKa = 10.66PPLYY256 pKa = 8.45PQPDD260 pKa = 3.64ADD262 pKa = 3.79STTFGASSGYY272 pKa = 10.03GFSPVAPGEE281 pKa = 4.14YY282 pKa = 10.16ASDD285 pKa = 3.9YY286 pKa = 10.71NDD288 pKa = 3.7EE289 pKa = 4.23EE290 pKa = 5.05GEE292 pKa = 4.2YY293 pKa = 10.97ADD295 pKa = 4.73YY296 pKa = 11.45DD297 pKa = 3.88GGNPNQDD304 pKa = 2.63EE305 pKa = 4.48SAAIPAEE312 pKa = 4.19TTAPSGQANQGGAIPGYY329 pKa = 9.38GFPAGSPTEE338 pKa = 4.46SMAADD343 pKa = 3.95MPDD346 pKa = 3.02VDD348 pKa = 4.31LTEE351 pKa = 5.85DD352 pKa = 3.69DD353 pKa = 4.85GEE355 pKa = 4.67TEE357 pKa = 4.18AEE359 pKa = 4.11LPGEE363 pKa = 4.13IGLIEE368 pKa = 4.2TEE370 pKa = 4.23EE371 pKa = 3.96ATGFDD376 pKa = 4.9DD377 pKa = 5.89FGEE380 pKa = 4.54TPAAEE385 pKa = 4.2MAPHH389 pKa = 7.02EE390 pKa = 4.85DD391 pKa = 3.74AFPPPTDD398 pKa = 3.35TPTPNPYY405 pKa = 10.19PMEE408 pKa = 5.1SGGYY412 pKa = 9.99GNAPDD417 pKa = 5.03PGFEE421 pKa = 5.11AYY423 pKa = 9.19GQPEE427 pKa = 3.63AWGNDD432 pKa = 2.89GSAADD437 pKa = 4.25TTPGGYY443 pKa = 9.47PEE445 pKa = 4.63VPRR448 pKa = 11.84DD449 pKa = 3.22PNFAYY454 pKa = 10.02GYY456 pKa = 8.3AQPGAPAGDD465 pKa = 3.63YY466 pKa = 10.78GYY468 pKa = 10.6QEE470 pKa = 3.77QDD472 pKa = 2.44GVYY475 pKa = 10.33AEE477 pKa = 4.91NSGEE481 pKa = 4.45TNWEE485 pKa = 3.99AMPQPGNDD493 pKa = 3.15TPMPQGNNPYY503 pKa = 9.56DD504 pKa = 3.9APGSAQFEE512 pKa = 4.14AAGYY516 pKa = 9.65GDD518 pKa = 4.97AYY520 pKa = 10.75ANEE523 pKa = 4.12AATGYY528 pKa = 8.38EE529 pKa = 4.24ANFGEE534 pKa = 5.07DD535 pKa = 4.16GYY537 pKa = 9.59TEE539 pKa = 4.06TGMAAAGGEE548 pKa = 4.75TNGEE552 pKa = 4.25TYY554 pKa = 10.73NPSEE558 pKa = 4.13AGQYY562 pKa = 9.74PEE564 pKa = 4.07YY565 pKa = 10.78ADD567 pKa = 3.91YY568 pKa = 11.28PEE570 pKa = 4.6TPGTNPDD577 pKa = 3.03VSEE580 pKa = 4.51FAVEE584 pKa = 4.57TPFTPTDD591 pKa = 3.36MSNGYY596 pKa = 7.55QQYY599 pKa = 10.35DD600 pKa = 3.81AYY602 pKa = 9.4PEE604 pKa = 4.02QVSPEE609 pKa = 3.94NGGTGEE615 pKa = 4.03FAAGFGGDD623 pKa = 3.91EE624 pKa = 4.82GYY626 pKa = 11.08SDD628 pKa = 3.57ATGYY632 pKa = 11.18DD633 pKa = 3.74DD634 pKa = 6.1DD635 pKa = 4.35NAYY638 pKa = 8.27PTEE641 pKa = 4.17TAAGMGDD648 pKa = 3.17AGGGDD653 pKa = 4.61FEE655 pKa = 4.53ATGYY659 pKa = 10.22PEE661 pKa = 3.93TNAEE665 pKa = 4.08FTEE668 pKa = 4.41DD669 pKa = 2.97TGGFATPGWGTEE681 pKa = 4.32TYY683 pKa = 10.32PGTEE687 pKa = 4.45PDD689 pKa = 3.3TLAAGSADD697 pKa = 3.8GLDD700 pKa = 3.83SPAGGTTATAAVAATTPAFNLPGEE724 pKa = 4.58APAVPPVDD732 pKa = 3.75TPVEE736 pKa = 4.31PIRR739 pKa = 11.84NPFTPATGNAAGAVEE754 pKa = 4.31SSGIAMPPHH763 pKa = 6.36EE764 pKa = 5.34PLTEE768 pKa = 4.13VVTPDD773 pKa = 3.76LPTPGVGLDD782 pKa = 3.6LSSLKK787 pKa = 8.42TTDD790 pKa = 3.62SKK792 pKa = 10.34TEE794 pKa = 4.0PYY796 pKa = 10.2SAGEE800 pKa = 4.15APSGQAPHH808 pKa = 6.9EE809 pKa = 4.61PGEE812 pKa = 4.36TNATGKK818 pKa = 10.32FSAAALEE825 pKa = 4.56ALDD828 pKa = 4.56QARR831 pKa = 11.84NDD833 pKa = 3.84PDD835 pKa = 3.52LGKK838 pKa = 10.64ASLTNSVAAANQVASTASPNHH859 pKa = 5.78KK860 pKa = 9.91RR861 pKa = 11.84FAEE864 pKa = 4.32VTSRR868 pKa = 11.84ATEE871 pKa = 4.09TAAEE875 pKa = 4.15TATVEE880 pKa = 4.12NGRR883 pKa = 11.84PDD885 pKa = 3.15ATQPPKK891 pKa = 10.84NLARR895 pKa = 11.84NRR897 pKa = 11.84GDD899 pKa = 3.31KK900 pKa = 10.66PGGGDD905 pKa = 3.44TVTAAYY911 pKa = 7.74PAGLAGIPGLEE922 pKa = 4.08PLNPAEE928 pKa = 4.99LLTPATGIEE937 pKa = 4.71TVTGPTFTEE946 pKa = 4.25VPIDD950 pKa = 3.56HH951 pKa = 6.85EE952 pKa = 4.32AASRR956 pKa = 11.84LAEE959 pKa = 4.09TTDD962 pKa = 2.93IDD964 pKa = 4.26TADD967 pKa = 3.79EE968 pKa = 4.2TASTPRR974 pKa = 11.84EE975 pKa = 4.06TEE977 pKa = 3.87EE978 pKa = 3.92PSTPKK983 pKa = 10.21EE984 pKa = 4.07IPEE987 pKa = 4.13PTPKK991 pKa = 10.27EE992 pKa = 4.02IPEE995 pKa = 4.09PTPKK999 pKa = 9.42EE1000 pKa = 4.46TEE1002 pKa = 3.81TSTFSGTKK1010 pKa = 10.12LPEE1013 pKa = 4.24NANPRR1018 pKa = 11.84PLLQGEE1024 pKa = 4.45EE1025 pKa = 4.22PFGEE1029 pKa = 4.32VTEE1032 pKa = 4.39EE1033 pKa = 4.18GRR1035 pKa = 11.84FVPHH1039 pKa = 6.52FLAGYY1044 pKa = 7.56RR1045 pKa = 11.84TNLDD1049 pKa = 3.81TFTSAYY1055 pKa = 10.15GSLEE1059 pKa = 4.08EE1060 pKa = 5.09HH1061 pKa = 6.25SQPHH1065 pKa = 5.54AKK1067 pKa = 10.09KK1068 pKa = 9.7PEE1070 pKa = 4.47SIAKK1074 pKa = 10.23LEE1076 pKa = 4.12AEE1078 pKa = 4.46QPRR1081 pKa = 11.84KK1082 pKa = 10.48SITTAYY1088 pKa = 9.9LVTVTTSRR1096 pKa = 11.84PLGSDD1101 pKa = 2.9DD1102 pKa = 3.59AVFVRR1107 pKa = 11.84FPDD1110 pKa = 3.75GRR1112 pKa = 11.84EE1113 pKa = 3.93LPVTSAYY1120 pKa = 11.02SDD1122 pKa = 3.66FNGVHH1127 pKa = 6.36LNIPRR1132 pKa = 11.84LL1133 pKa = 3.66
MM1 pKa = 8.04SEE3 pKa = 4.45EE4 pKa = 4.1YY5 pKa = 10.16TPQLLTSEE13 pKa = 5.17DD14 pKa = 3.94VFTVQFPATKK24 pKa = 10.14FRR26 pKa = 11.84DD27 pKa = 4.43GYY29 pKa = 10.67DD30 pKa = 3.03QNQIDD35 pKa = 4.67DD36 pKa = 4.11YY37 pKa = 10.39LDD39 pKa = 3.29EE40 pKa = 4.38VVRR43 pKa = 11.84VLSYY47 pKa = 11.4YY48 pKa = 9.85EE49 pKa = 4.31ALSAAPEE56 pKa = 4.22AEE58 pKa = 3.8VDD60 pKa = 3.41LTYY63 pKa = 9.95ITVRR67 pKa = 11.84GVDD70 pKa = 3.47VRR72 pKa = 11.84EE73 pKa = 3.62VDD75 pKa = 3.92FDD77 pKa = 3.77YY78 pKa = 11.3TRR80 pKa = 11.84MRR82 pKa = 11.84VGYY85 pKa = 9.8DD86 pKa = 2.92QDD88 pKa = 4.8AVDD91 pKa = 5.52DD92 pKa = 4.27YY93 pKa = 11.24LDD95 pKa = 3.46QVAATLEE102 pKa = 4.35AYY104 pKa = 9.08EE105 pKa = 4.16NAYY108 pKa = 9.96GIPPSDD114 pKa = 3.18QRR116 pKa = 11.84YY117 pKa = 8.28QVLQVDD123 pKa = 5.36GPALEE128 pKa = 5.45AEE130 pKa = 4.7AAQQAAAAGEE140 pKa = 4.2LPAEE144 pKa = 4.37VSAPVALGDD153 pKa = 4.09SAAGGAPVTAGNMLSLGASVGGAVPLDD180 pKa = 4.1AAPSAPDD187 pKa = 3.19KK188 pKa = 10.87AASLGASEE196 pKa = 4.63TAPNSPGYY204 pKa = 10.39SSVLGTPPARR214 pKa = 11.84QPGVGEE220 pKa = 4.17FNEE223 pKa = 4.33APQYY227 pKa = 10.84PGDD230 pKa = 4.0SLDD233 pKa = 3.15WMQAPGQSPASSPQNYY249 pKa = 8.69PDD251 pKa = 4.1YY252 pKa = 10.66PPLYY256 pKa = 8.45PQPDD260 pKa = 3.64ADD262 pKa = 3.79STTFGASSGYY272 pKa = 10.03GFSPVAPGEE281 pKa = 4.14YY282 pKa = 10.16ASDD285 pKa = 3.9YY286 pKa = 10.71NDD288 pKa = 3.7EE289 pKa = 4.23EE290 pKa = 5.05GEE292 pKa = 4.2YY293 pKa = 10.97ADD295 pKa = 4.73YY296 pKa = 11.45DD297 pKa = 3.88GGNPNQDD304 pKa = 2.63EE305 pKa = 4.48SAAIPAEE312 pKa = 4.19TTAPSGQANQGGAIPGYY329 pKa = 9.38GFPAGSPTEE338 pKa = 4.46SMAADD343 pKa = 3.95MPDD346 pKa = 3.02VDD348 pKa = 4.31LTEE351 pKa = 5.85DD352 pKa = 3.69DD353 pKa = 4.85GEE355 pKa = 4.67TEE357 pKa = 4.18AEE359 pKa = 4.11LPGEE363 pKa = 4.13IGLIEE368 pKa = 4.2TEE370 pKa = 4.23EE371 pKa = 3.96ATGFDD376 pKa = 4.9DD377 pKa = 5.89FGEE380 pKa = 4.54TPAAEE385 pKa = 4.2MAPHH389 pKa = 7.02EE390 pKa = 4.85DD391 pKa = 3.74AFPPPTDD398 pKa = 3.35TPTPNPYY405 pKa = 10.19PMEE408 pKa = 5.1SGGYY412 pKa = 9.99GNAPDD417 pKa = 5.03PGFEE421 pKa = 5.11AYY423 pKa = 9.19GQPEE427 pKa = 3.63AWGNDD432 pKa = 2.89GSAADD437 pKa = 4.25TTPGGYY443 pKa = 9.47PEE445 pKa = 4.63VPRR448 pKa = 11.84DD449 pKa = 3.22PNFAYY454 pKa = 10.02GYY456 pKa = 8.3AQPGAPAGDD465 pKa = 3.63YY466 pKa = 10.78GYY468 pKa = 10.6QEE470 pKa = 3.77QDD472 pKa = 2.44GVYY475 pKa = 10.33AEE477 pKa = 4.91NSGEE481 pKa = 4.45TNWEE485 pKa = 3.99AMPQPGNDD493 pKa = 3.15TPMPQGNNPYY503 pKa = 9.56DD504 pKa = 3.9APGSAQFEE512 pKa = 4.14AAGYY516 pKa = 9.65GDD518 pKa = 4.97AYY520 pKa = 10.75ANEE523 pKa = 4.12AATGYY528 pKa = 8.38EE529 pKa = 4.24ANFGEE534 pKa = 5.07DD535 pKa = 4.16GYY537 pKa = 9.59TEE539 pKa = 4.06TGMAAAGGEE548 pKa = 4.75TNGEE552 pKa = 4.25TYY554 pKa = 10.73NPSEE558 pKa = 4.13AGQYY562 pKa = 9.74PEE564 pKa = 4.07YY565 pKa = 10.78ADD567 pKa = 3.91YY568 pKa = 11.28PEE570 pKa = 4.6TPGTNPDD577 pKa = 3.03VSEE580 pKa = 4.51FAVEE584 pKa = 4.57TPFTPTDD591 pKa = 3.36MSNGYY596 pKa = 7.55QQYY599 pKa = 10.35DD600 pKa = 3.81AYY602 pKa = 9.4PEE604 pKa = 4.02QVSPEE609 pKa = 3.94NGGTGEE615 pKa = 4.03FAAGFGGDD623 pKa = 3.91EE624 pKa = 4.82GYY626 pKa = 11.08SDD628 pKa = 3.57ATGYY632 pKa = 11.18DD633 pKa = 3.74DD634 pKa = 6.1DD635 pKa = 4.35NAYY638 pKa = 8.27PTEE641 pKa = 4.17TAAGMGDD648 pKa = 3.17AGGGDD653 pKa = 4.61FEE655 pKa = 4.53ATGYY659 pKa = 10.22PEE661 pKa = 3.93TNAEE665 pKa = 4.08FTEE668 pKa = 4.41DD669 pKa = 2.97TGGFATPGWGTEE681 pKa = 4.32TYY683 pKa = 10.32PGTEE687 pKa = 4.45PDD689 pKa = 3.3TLAAGSADD697 pKa = 3.8GLDD700 pKa = 3.83SPAGGTTATAAVAATTPAFNLPGEE724 pKa = 4.58APAVPPVDD732 pKa = 3.75TPVEE736 pKa = 4.31PIRR739 pKa = 11.84NPFTPATGNAAGAVEE754 pKa = 4.31SSGIAMPPHH763 pKa = 6.36EE764 pKa = 5.34PLTEE768 pKa = 4.13VVTPDD773 pKa = 3.76LPTPGVGLDD782 pKa = 3.6LSSLKK787 pKa = 8.42TTDD790 pKa = 3.62SKK792 pKa = 10.34TEE794 pKa = 4.0PYY796 pKa = 10.2SAGEE800 pKa = 4.15APSGQAPHH808 pKa = 6.9EE809 pKa = 4.61PGEE812 pKa = 4.36TNATGKK818 pKa = 10.32FSAAALEE825 pKa = 4.56ALDD828 pKa = 4.56QARR831 pKa = 11.84NDD833 pKa = 3.84PDD835 pKa = 3.52LGKK838 pKa = 10.64ASLTNSVAAANQVASTASPNHH859 pKa = 5.78KK860 pKa = 9.91RR861 pKa = 11.84FAEE864 pKa = 4.32VTSRR868 pKa = 11.84ATEE871 pKa = 4.09TAAEE875 pKa = 4.15TATVEE880 pKa = 4.12NGRR883 pKa = 11.84PDD885 pKa = 3.15ATQPPKK891 pKa = 10.84NLARR895 pKa = 11.84NRR897 pKa = 11.84GDD899 pKa = 3.31KK900 pKa = 10.66PGGGDD905 pKa = 3.44TVTAAYY911 pKa = 7.74PAGLAGIPGLEE922 pKa = 4.08PLNPAEE928 pKa = 4.99LLTPATGIEE937 pKa = 4.71TVTGPTFTEE946 pKa = 4.25VPIDD950 pKa = 3.56HH951 pKa = 6.85EE952 pKa = 4.32AASRR956 pKa = 11.84LAEE959 pKa = 4.09TTDD962 pKa = 2.93IDD964 pKa = 4.26TADD967 pKa = 3.79EE968 pKa = 4.2TASTPRR974 pKa = 11.84EE975 pKa = 4.06TEE977 pKa = 3.87EE978 pKa = 3.92PSTPKK983 pKa = 10.21EE984 pKa = 4.07IPEE987 pKa = 4.13PTPKK991 pKa = 10.27EE992 pKa = 4.02IPEE995 pKa = 4.09PTPKK999 pKa = 9.42EE1000 pKa = 4.46TEE1002 pKa = 3.81TSTFSGTKK1010 pKa = 10.12LPEE1013 pKa = 4.24NANPRR1018 pKa = 11.84PLLQGEE1024 pKa = 4.45EE1025 pKa = 4.22PFGEE1029 pKa = 4.32VTEE1032 pKa = 4.39EE1033 pKa = 4.18GRR1035 pKa = 11.84FVPHH1039 pKa = 6.52FLAGYY1044 pKa = 7.56RR1045 pKa = 11.84TNLDD1049 pKa = 3.81TFTSAYY1055 pKa = 10.15GSLEE1059 pKa = 4.08EE1060 pKa = 5.09HH1061 pKa = 6.25SQPHH1065 pKa = 5.54AKK1067 pKa = 10.09KK1068 pKa = 9.7PEE1070 pKa = 4.47SIAKK1074 pKa = 10.23LEE1076 pKa = 4.12AEE1078 pKa = 4.46QPRR1081 pKa = 11.84KK1082 pKa = 10.48SITTAYY1088 pKa = 9.9LVTVTTSRR1096 pKa = 11.84PLGSDD1101 pKa = 2.9DD1102 pKa = 3.59AVFVRR1107 pKa = 11.84FPDD1110 pKa = 3.75GRR1112 pKa = 11.84EE1113 pKa = 3.93LPVTSAYY1120 pKa = 11.02SDD1122 pKa = 3.66FNGVHH1127 pKa = 6.36LNIPRR1132 pKa = 11.84LL1133 pKa = 3.66
Molecular weight: 117.73 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E0QN63|E0QN63_9ACTO Glycosyltransferase group 2 family protein OS=Mobiluncus mulieris ATCC 35239 OX=871571 GN=rfaG PE=4 SV=1
MM1 pKa = 7.62TKK3 pKa = 9.14RR4 pKa = 11.84TYY6 pKa = 10.01QPHH9 pKa = 5.09NRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 10.24KK16 pKa = 8.45HH17 pKa = 4.25GFRR20 pKa = 11.84NRR22 pKa = 11.84MATRR26 pKa = 11.84AGRR29 pKa = 11.84AVISARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84GRR40 pKa = 11.84KK41 pKa = 8.5QLAVV45 pKa = 3.36
MM1 pKa = 7.62TKK3 pKa = 9.14RR4 pKa = 11.84TYY6 pKa = 10.01QPHH9 pKa = 5.09NRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 10.24KK16 pKa = 8.45HH17 pKa = 4.25GFRR20 pKa = 11.84NRR22 pKa = 11.84MATRR26 pKa = 11.84AGRR29 pKa = 11.84AVISARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84GRR40 pKa = 11.84KK41 pKa = 8.5QLAVV45 pKa = 3.36
Molecular weight: 5.4 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
730285 |
37 |
7651 |
307.0 |
33.38 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.052 ± 0.062 | 0.876 ± 0.016 |
5.253 ± 0.043 | 5.915 ± 0.049 |
3.421 ± 0.034 | 8.223 ± 0.054 |
1.973 ± 0.023 | 5.022 ± 0.047 |
4.28 ± 0.078 | 9.844 ± 0.084 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.269 ± 0.029 | 3.286 ± 0.041 |
5.157 ± 0.048 | 3.693 ± 0.028 |
5.88 ± 0.063 | 6.032 ± 0.041 |
6.126 ± 0.055 | 7.847 ± 0.048 |
1.439 ± 0.024 | 2.41 ± 0.033 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
