
Synechococcus phage S-CBP1
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Autographiviridae; unclassified Autographiviridae
Average proteome isoelectric point is 6.1
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 50 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
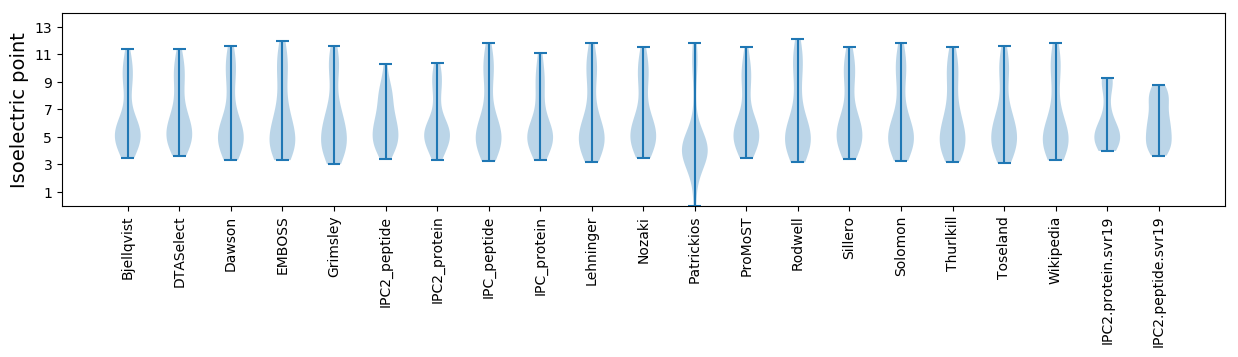
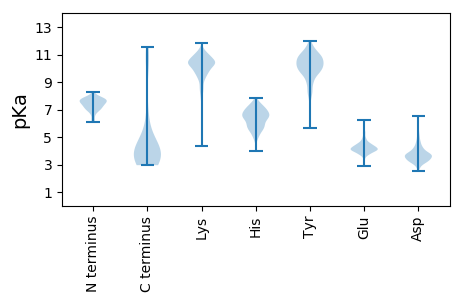
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A096VKD1|A0A096VKD1_9CAUD Uncharacterized protein OS=Synechococcus phage S-CBP1 OX=1273711 GN=S-CBP1_0008 PE=4 SV=1
MM1 pKa = 6.97TTTIPIVLTGQSLVDD16 pKa = 3.6YY17 pKa = 10.39VDD19 pKa = 3.2EE20 pKa = 4.57RR21 pKa = 11.84KK22 pKa = 10.24ALIEE26 pKa = 4.27RR27 pKa = 11.84GEE29 pKa = 4.07LTRR32 pKa = 11.84TDD34 pKa = 4.64LIKK37 pKa = 10.96DD38 pKa = 3.26AGYY41 pKa = 11.23VSDD44 pKa = 4.09NGKK47 pKa = 9.94ARR49 pKa = 11.84YY50 pKa = 9.32VDD52 pKa = 5.86FYY54 pKa = 11.4TEE56 pKa = 4.04LLNARR61 pKa = 11.84GITPVTNRR69 pKa = 11.84DD70 pKa = 3.35IEE72 pKa = 4.55DD73 pKa = 3.71NEE75 pKa = 3.87YY76 pKa = 11.16DD77 pKa = 4.81ALNEE81 pKa = 4.16DD82 pKa = 3.79EE83 pKa = 4.93KK84 pKa = 11.67ALYY87 pKa = 10.14DD88 pKa = 3.92YY89 pKa = 11.32LDD91 pKa = 3.84EE92 pKa = 5.04QSFTEE97 pKa = 4.15KK98 pKa = 9.38WDD100 pKa = 3.39HH101 pKa = 6.38EE102 pKa = 4.36EE103 pKa = 4.29FIEE106 pKa = 5.32FLVEE110 pKa = 4.17LKK112 pKa = 11.03DD113 pKa = 3.69NGIEE117 pKa = 4.27TVSSFEE123 pKa = 4.33DD124 pKa = 3.03AFYY127 pKa = 10.85SYY129 pKa = 11.43NDD131 pKa = 3.58EE132 pKa = 4.26YY133 pKa = 10.38WAEE136 pKa = 3.73RR137 pKa = 11.84HH138 pKa = 5.44FAEE141 pKa = 4.95EE142 pKa = 4.16YY143 pKa = 8.85VNEE146 pKa = 4.21VEE148 pKa = 5.33SIQDD152 pKa = 3.62SIVYY156 pKa = 9.31HH157 pKa = 6.66AIDD160 pKa = 3.48WQAVWDD166 pKa = 3.85HH167 pKa = 4.94QLRR170 pKa = 11.84YY171 pKa = 10.14DD172 pKa = 3.96FCTIEE177 pKa = 3.91HH178 pKa = 6.53GGWTFFFRR186 pKa = 11.84NYY188 pKa = 10.46
MM1 pKa = 6.97TTTIPIVLTGQSLVDD16 pKa = 3.6YY17 pKa = 10.39VDD19 pKa = 3.2EE20 pKa = 4.57RR21 pKa = 11.84KK22 pKa = 10.24ALIEE26 pKa = 4.27RR27 pKa = 11.84GEE29 pKa = 4.07LTRR32 pKa = 11.84TDD34 pKa = 4.64LIKK37 pKa = 10.96DD38 pKa = 3.26AGYY41 pKa = 11.23VSDD44 pKa = 4.09NGKK47 pKa = 9.94ARR49 pKa = 11.84YY50 pKa = 9.32VDD52 pKa = 5.86FYY54 pKa = 11.4TEE56 pKa = 4.04LLNARR61 pKa = 11.84GITPVTNRR69 pKa = 11.84DD70 pKa = 3.35IEE72 pKa = 4.55DD73 pKa = 3.71NEE75 pKa = 3.87YY76 pKa = 11.16DD77 pKa = 4.81ALNEE81 pKa = 4.16DD82 pKa = 3.79EE83 pKa = 4.93KK84 pKa = 11.67ALYY87 pKa = 10.14DD88 pKa = 3.92YY89 pKa = 11.32LDD91 pKa = 3.84EE92 pKa = 5.04QSFTEE97 pKa = 4.15KK98 pKa = 9.38WDD100 pKa = 3.39HH101 pKa = 6.38EE102 pKa = 4.36EE103 pKa = 4.29FIEE106 pKa = 5.32FLVEE110 pKa = 4.17LKK112 pKa = 11.03DD113 pKa = 3.69NGIEE117 pKa = 4.27TVSSFEE123 pKa = 4.33DD124 pKa = 3.03AFYY127 pKa = 10.85SYY129 pKa = 11.43NDD131 pKa = 3.58EE132 pKa = 4.26YY133 pKa = 10.38WAEE136 pKa = 3.73RR137 pKa = 11.84HH138 pKa = 5.44FAEE141 pKa = 4.95EE142 pKa = 4.16YY143 pKa = 8.85VNEE146 pKa = 4.21VEE148 pKa = 5.33SIQDD152 pKa = 3.62SIVYY156 pKa = 9.31HH157 pKa = 6.66AIDD160 pKa = 3.48WQAVWDD166 pKa = 3.85HH167 pKa = 4.94QLRR170 pKa = 11.84YY171 pKa = 10.14DD172 pKa = 3.96FCTIEE177 pKa = 3.91HH178 pKa = 6.53GGWTFFFRR186 pKa = 11.84NYY188 pKa = 10.46
Molecular weight: 22.35 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A096VKE7|A0A096VKE7_9CAUD Uncharacterized protein OS=Synechococcus phage S-CBP1 OX=1273711 GN=S-CBP1_0023 PE=4 SV=1
MM1 pKa = 6.26TRR3 pKa = 11.84ARR5 pKa = 11.84IEE7 pKa = 3.97YY8 pKa = 10.12ARR10 pKa = 11.84GRR12 pKa = 11.84GGGGRR17 pKa = 11.84GGGGRR22 pKa = 11.84GGGSRR27 pKa = 11.84GRR29 pKa = 11.84GGGGGGRR36 pKa = 11.84SSGGGGGGRR45 pKa = 11.84SSGGGGGGGRR55 pKa = 11.84AAASSAGRR63 pKa = 11.84SAASTGSFTSANAQALRR80 pKa = 11.84QAGVSRR86 pKa = 11.84SRR88 pKa = 11.84IQNLRR93 pKa = 11.84SQSQNINNRR102 pKa = 11.84NSAAADD108 pKa = 3.04ARR110 pKa = 11.84AAMDD114 pKa = 4.22PSVAGAMEE122 pKa = 4.12AQANRR127 pKa = 11.84GLNIGQQFYY136 pKa = 10.96DD137 pKa = 3.46SPEE140 pKa = 3.86FQQALGQSGLSPQQIAQYY158 pKa = 11.33AMSQGYY164 pKa = 8.9GLGNQFMDD172 pKa = 3.64QFGTTTTNRR181 pKa = 11.84SISGATKK188 pKa = 9.77YY189 pKa = 10.41RR190 pKa = 11.84QALKK194 pKa = 10.19IAAAQGRR201 pKa = 11.84GTNLIGGRR209 pKa = 11.84EE210 pKa = 4.05AKK212 pKa = 10.41QIAKK216 pKa = 9.3QFKK219 pKa = 10.29NKK221 pKa = 9.12TDD223 pKa = 3.81LNFARR228 pKa = 11.84SLDD231 pKa = 3.88KK232 pKa = 11.18INANKK237 pKa = 9.43PNMMPIGLTGGVLNRR252 pKa = 11.84LSRR255 pKa = 11.84NKK257 pKa = 10.43DD258 pKa = 2.99GSLSRR263 pKa = 11.84MGMMPSSDD271 pKa = 3.62GFLKK275 pKa = 10.52ILANMNDD282 pKa = 3.13RR283 pKa = 11.84WTAGDD288 pKa = 3.72ARR290 pKa = 11.84NGNPSKK296 pKa = 10.75RR297 pKa = 11.84IPGTGKK303 pKa = 9.69FRR305 pKa = 11.84RR306 pKa = 11.84TDD308 pKa = 3.48TIYY311 pKa = 11.32GLMSDD316 pKa = 3.76GSFRR320 pKa = 11.84QTPGVGKK327 pKa = 10.11GWADD331 pKa = 3.26TRR333 pKa = 11.84FGQPEE338 pKa = 4.15TTQYY342 pKa = 10.17EE343 pKa = 4.29PWYY346 pKa = 9.04TPPEE350 pKa = 4.36DD351 pKa = 3.29QGVASVEE358 pKa = 4.4DD359 pKa = 4.07YY360 pKa = 11.65GEE362 pKa = 4.43APLPPEE368 pKa = 4.4EE369 pKa = 5.33PMTTDD374 pKa = 3.41SNQSGAGADD383 pKa = 3.53LASFATGWKK392 pKa = 8.36GARR395 pKa = 11.84GARR398 pKa = 11.84ARR400 pKa = 11.84AGRR403 pKa = 11.84KK404 pKa = 8.64AQGTGSMRR412 pKa = 11.84IAPTYY417 pKa = 9.38GAGVGSNFGG426 pKa = 3.39
MM1 pKa = 6.26TRR3 pKa = 11.84ARR5 pKa = 11.84IEE7 pKa = 3.97YY8 pKa = 10.12ARR10 pKa = 11.84GRR12 pKa = 11.84GGGGRR17 pKa = 11.84GGGGRR22 pKa = 11.84GGGSRR27 pKa = 11.84GRR29 pKa = 11.84GGGGGGRR36 pKa = 11.84SSGGGGGGRR45 pKa = 11.84SSGGGGGGGRR55 pKa = 11.84AAASSAGRR63 pKa = 11.84SAASTGSFTSANAQALRR80 pKa = 11.84QAGVSRR86 pKa = 11.84SRR88 pKa = 11.84IQNLRR93 pKa = 11.84SQSQNINNRR102 pKa = 11.84NSAAADD108 pKa = 3.04ARR110 pKa = 11.84AAMDD114 pKa = 4.22PSVAGAMEE122 pKa = 4.12AQANRR127 pKa = 11.84GLNIGQQFYY136 pKa = 10.96DD137 pKa = 3.46SPEE140 pKa = 3.86FQQALGQSGLSPQQIAQYY158 pKa = 11.33AMSQGYY164 pKa = 8.9GLGNQFMDD172 pKa = 3.64QFGTTTTNRR181 pKa = 11.84SISGATKK188 pKa = 9.77YY189 pKa = 10.41RR190 pKa = 11.84QALKK194 pKa = 10.19IAAAQGRR201 pKa = 11.84GTNLIGGRR209 pKa = 11.84EE210 pKa = 4.05AKK212 pKa = 10.41QIAKK216 pKa = 9.3QFKK219 pKa = 10.29NKK221 pKa = 9.12TDD223 pKa = 3.81LNFARR228 pKa = 11.84SLDD231 pKa = 3.88KK232 pKa = 11.18INANKK237 pKa = 9.43PNMMPIGLTGGVLNRR252 pKa = 11.84LSRR255 pKa = 11.84NKK257 pKa = 10.43DD258 pKa = 2.99GSLSRR263 pKa = 11.84MGMMPSSDD271 pKa = 3.62GFLKK275 pKa = 10.52ILANMNDD282 pKa = 3.13RR283 pKa = 11.84WTAGDD288 pKa = 3.72ARR290 pKa = 11.84NGNPSKK296 pKa = 10.75RR297 pKa = 11.84IPGTGKK303 pKa = 9.69FRR305 pKa = 11.84RR306 pKa = 11.84TDD308 pKa = 3.48TIYY311 pKa = 11.32GLMSDD316 pKa = 3.76GSFRR320 pKa = 11.84QTPGVGKK327 pKa = 10.11GWADD331 pKa = 3.26TRR333 pKa = 11.84FGQPEE338 pKa = 4.15TTQYY342 pKa = 10.17EE343 pKa = 4.29PWYY346 pKa = 9.04TPPEE350 pKa = 4.36DD351 pKa = 3.29QGVASVEE358 pKa = 4.4DD359 pKa = 4.07YY360 pKa = 11.65GEE362 pKa = 4.43APLPPEE368 pKa = 4.4EE369 pKa = 5.33PMTTDD374 pKa = 3.41SNQSGAGADD383 pKa = 3.53LASFATGWKK392 pKa = 8.36GARR395 pKa = 11.84GARR398 pKa = 11.84ARR400 pKa = 11.84AGRR403 pKa = 11.84KK404 pKa = 8.64AQGTGSMRR412 pKa = 11.84IAPTYY417 pKa = 9.38GAGVGSNFGG426 pKa = 3.39
Molecular weight: 43.99 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
13376 |
32 |
1402 |
267.5 |
29.55 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.824 ± 0.621 | 0.733 ± 0.159 |
6.347 ± 0.3 | 6.011 ± 0.422 |
3.514 ± 0.186 | 7.401 ± 0.447 |
1.391 ± 0.185 | 5.084 ± 0.208 |
5.233 ± 0.457 | 8.074 ± 0.348 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.385 ± 0.197 | 4.987 ± 0.313 |
4.224 ± 0.229 | 4.575 ± 0.296 |
5.51 ± 0.351 | 6.788 ± 0.463 |
6.699 ± 0.596 | 6.28 ± 0.344 |
1.368 ± 0.147 | 3.574 ± 0.267 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
