
Laurel Lake virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Ellioviricetes; Bunyavirales; Phenuiviridae; Laulavirus; Laurel Lake laulavirus
Average proteome isoelectric point is 6.79
Get precalculated fractions of proteins
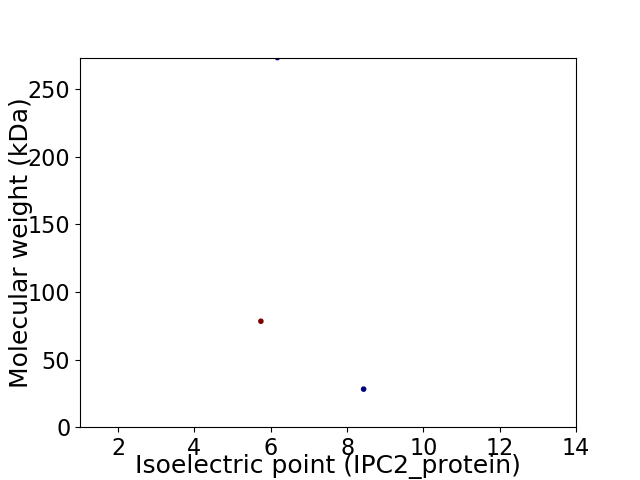
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A2K9YNI6|A0A2K9YNI6_9VIRU ORF1 OS=Laurel Lake virus OX=2027354 PE=4 SV=1
MM1 pKa = 7.72SNQTNMTDD9 pKa = 3.21VTSDD13 pKa = 3.49YY14 pKa = 11.44LNLPRR19 pKa = 11.84EE20 pKa = 4.18LQSIVEE26 pKa = 4.04NNILEE31 pKa = 4.42AAGEE35 pKa = 4.13EE36 pKa = 4.18FLQSIKK42 pKa = 10.49LASEE46 pKa = 4.15LRR48 pKa = 11.84RR49 pKa = 11.84DD50 pKa = 4.24LEE52 pKa = 4.72SSTQPDD58 pKa = 3.42DD59 pKa = 3.72DD60 pKa = 3.91RR61 pKa = 11.84RR62 pKa = 11.84NLRR65 pKa = 11.84LIRR68 pKa = 11.84GRR70 pKa = 11.84LGMSLLTSSLEE81 pKa = 4.17CPDD84 pKa = 4.53DD85 pKa = 4.28SFISSMGIGYY95 pKa = 10.07CYY97 pKa = 10.41LNSITQEE104 pKa = 3.83FRR106 pKa = 11.84IKK108 pKa = 10.44ALKK111 pKa = 7.67THH113 pKa = 5.87YY114 pKa = 10.03VWPTLQEE121 pKa = 3.94LFSRR125 pKa = 11.84WEE127 pKa = 3.73THH129 pKa = 4.67MVSGFRR135 pKa = 11.84LYY137 pKa = 9.78KK138 pKa = 9.92TVLSGKK144 pKa = 8.54LTVLHH149 pKa = 6.66LSYY152 pKa = 10.92SAVKK156 pKa = 9.9GVEE159 pKa = 4.03FVPIEE164 pKa = 3.97MQNQCNYY171 pKa = 8.72CHH173 pKa = 6.63AVSKK177 pKa = 10.73SYY179 pKa = 8.86TWLRR183 pKa = 11.84YY184 pKa = 8.85PKK186 pKa = 9.28LTHH189 pKa = 7.34DD190 pKa = 4.49VNCKK194 pKa = 8.71HH195 pKa = 5.57TVKK198 pKa = 10.81DD199 pKa = 4.63LISTPLDD206 pKa = 3.37SMYY209 pKa = 11.22LEE211 pKa = 4.69LKK213 pKa = 10.67VGGLFSRR220 pKa = 11.84LRR222 pKa = 11.84SPSQSTPTTSRR233 pKa = 11.84KK234 pKa = 8.99RR235 pKa = 11.84SKK237 pKa = 10.88SITDD241 pKa = 3.53SKK243 pKa = 10.86NHH245 pKa = 6.74EE246 pKa = 4.28DD247 pKa = 3.49RR248 pKa = 11.84QVIKK252 pKa = 10.46FDD254 pKa = 3.88RR255 pKa = 11.84SVKK258 pKa = 10.83GEE260 pKa = 3.92GKK262 pKa = 10.22EE263 pKa = 3.9PDD265 pKa = 3.48MSRR268 pKa = 11.84DD269 pKa = 3.58LYY271 pKa = 11.22SVPKK275 pKa = 7.78PTEE278 pKa = 3.71IEE280 pKa = 4.21TMSDD284 pKa = 2.71HH285 pKa = 7.0LNNHH289 pKa = 6.38KK290 pKa = 10.93SGDD293 pKa = 3.65TFPYY297 pKa = 8.9VSVVGTEE304 pKa = 4.75DD305 pKa = 3.14EE306 pKa = 4.6TKK308 pKa = 10.56DD309 pKa = 3.48GRR311 pKa = 11.84LSRR314 pKa = 11.84LRR316 pKa = 11.84AFRR319 pKa = 11.84SSYY322 pKa = 10.62GKK324 pKa = 9.92EE325 pKa = 3.99NKK327 pKa = 9.81PEE329 pKa = 3.66MRR331 pKa = 11.84INGVILDD338 pKa = 4.15EE339 pKa = 4.64LEE341 pKa = 4.11GTIKK345 pKa = 10.5VSNIGDD351 pKa = 3.77YY352 pKa = 10.99VNKK355 pKa = 10.42EE356 pKa = 4.05NLNKK360 pKa = 9.24QQRR363 pKa = 11.84SEE365 pKa = 4.1NKK367 pKa = 9.28RR368 pKa = 11.84FRR370 pKa = 11.84PLNEE374 pKa = 3.73ASFKK378 pKa = 10.89KK379 pKa = 10.11PYY381 pKa = 10.04LQIARR386 pKa = 11.84ITGEE390 pKa = 4.29YY391 pKa = 9.1VPLMSSTSDD400 pKa = 3.3YY401 pKa = 10.74TEE403 pKa = 5.1LYY405 pKa = 7.81FTLEE409 pKa = 4.43DD410 pKa = 4.84GRR412 pKa = 11.84LLDD415 pKa = 3.66NQVIIQSNKK424 pKa = 10.11LPTNQNGVFEE434 pKa = 5.05LSCDD438 pKa = 3.62YY439 pKa = 10.89CINLSDD445 pKa = 4.77INQLSLKK452 pKa = 9.72YY453 pKa = 10.04FLSRR457 pKa = 11.84PIMKK461 pKa = 10.3EE462 pKa = 3.73GFQWGAVSLTIRR474 pKa = 11.84VSEE477 pKa = 4.1SDD479 pKa = 3.52TPYY482 pKa = 9.9LTPKK486 pKa = 9.31VEE488 pKa = 3.77AMAIVRR494 pKa = 11.84IPFSTLEE501 pKa = 4.14EE502 pKa = 3.99QSKK505 pKa = 11.23DD506 pKa = 3.34PDD508 pKa = 3.58HH509 pKa = 7.08ADD511 pKa = 3.15VVFTAKK517 pKa = 10.21QVDD520 pKa = 3.93KK521 pKa = 11.18FKK523 pKa = 10.87EE524 pKa = 4.07LYY526 pKa = 10.02RR527 pKa = 11.84AGDD530 pKa = 3.42VMDD533 pKa = 4.0IDD535 pKa = 3.63EE536 pKa = 4.94AKK538 pKa = 10.42RR539 pKa = 11.84EE540 pKa = 3.78RR541 pKa = 11.84STINSYY547 pKa = 11.0SKK549 pKa = 9.02STIRR553 pKa = 11.84GVVKK557 pKa = 10.81GEE559 pKa = 4.24AGPSHH564 pKa = 7.42LGNQSGWEE572 pKa = 3.99HH573 pKa = 6.49LKK575 pKa = 11.18GMVKK579 pKa = 10.02PRR581 pKa = 11.84VEE583 pKa = 4.19EE584 pKa = 4.44GQASISVASGSEE596 pKa = 4.0EE597 pKa = 3.72VDD599 pKa = 2.56IDD601 pKa = 3.98VPSTTRR607 pKa = 11.84EE608 pKa = 3.98EE609 pKa = 4.39YY610 pKa = 10.39EE611 pKa = 4.06KK612 pKa = 10.6QQEE615 pKa = 4.25LLRR618 pKa = 11.84QQFQQTFSDD627 pKa = 3.72TTEE630 pKa = 4.28EE631 pKa = 4.15IPRR634 pKa = 11.84SEE636 pKa = 4.69LNRR639 pKa = 11.84SSSPINSDD647 pKa = 3.28PLSSVEE653 pKa = 3.97EE654 pKa = 4.2SSPPRR659 pKa = 11.84SPIKK663 pKa = 10.53SAMKK667 pKa = 9.81KK668 pKa = 9.79PRR670 pKa = 11.84FIEE673 pKa = 4.32DD674 pKa = 3.58TLKK677 pKa = 11.08KK678 pKa = 10.01PDD680 pKa = 3.66TSDD683 pKa = 3.34VYY685 pKa = 11.27QFNN688 pKa = 3.79
MM1 pKa = 7.72SNQTNMTDD9 pKa = 3.21VTSDD13 pKa = 3.49YY14 pKa = 11.44LNLPRR19 pKa = 11.84EE20 pKa = 4.18LQSIVEE26 pKa = 4.04NNILEE31 pKa = 4.42AAGEE35 pKa = 4.13EE36 pKa = 4.18FLQSIKK42 pKa = 10.49LASEE46 pKa = 4.15LRR48 pKa = 11.84RR49 pKa = 11.84DD50 pKa = 4.24LEE52 pKa = 4.72SSTQPDD58 pKa = 3.42DD59 pKa = 3.72DD60 pKa = 3.91RR61 pKa = 11.84RR62 pKa = 11.84NLRR65 pKa = 11.84LIRR68 pKa = 11.84GRR70 pKa = 11.84LGMSLLTSSLEE81 pKa = 4.17CPDD84 pKa = 4.53DD85 pKa = 4.28SFISSMGIGYY95 pKa = 10.07CYY97 pKa = 10.41LNSITQEE104 pKa = 3.83FRR106 pKa = 11.84IKK108 pKa = 10.44ALKK111 pKa = 7.67THH113 pKa = 5.87YY114 pKa = 10.03VWPTLQEE121 pKa = 3.94LFSRR125 pKa = 11.84WEE127 pKa = 3.73THH129 pKa = 4.67MVSGFRR135 pKa = 11.84LYY137 pKa = 9.78KK138 pKa = 9.92TVLSGKK144 pKa = 8.54LTVLHH149 pKa = 6.66LSYY152 pKa = 10.92SAVKK156 pKa = 9.9GVEE159 pKa = 4.03FVPIEE164 pKa = 3.97MQNQCNYY171 pKa = 8.72CHH173 pKa = 6.63AVSKK177 pKa = 10.73SYY179 pKa = 8.86TWLRR183 pKa = 11.84YY184 pKa = 8.85PKK186 pKa = 9.28LTHH189 pKa = 7.34DD190 pKa = 4.49VNCKK194 pKa = 8.71HH195 pKa = 5.57TVKK198 pKa = 10.81DD199 pKa = 4.63LISTPLDD206 pKa = 3.37SMYY209 pKa = 11.22LEE211 pKa = 4.69LKK213 pKa = 10.67VGGLFSRR220 pKa = 11.84LRR222 pKa = 11.84SPSQSTPTTSRR233 pKa = 11.84KK234 pKa = 8.99RR235 pKa = 11.84SKK237 pKa = 10.88SITDD241 pKa = 3.53SKK243 pKa = 10.86NHH245 pKa = 6.74EE246 pKa = 4.28DD247 pKa = 3.49RR248 pKa = 11.84QVIKK252 pKa = 10.46FDD254 pKa = 3.88RR255 pKa = 11.84SVKK258 pKa = 10.83GEE260 pKa = 3.92GKK262 pKa = 10.22EE263 pKa = 3.9PDD265 pKa = 3.48MSRR268 pKa = 11.84DD269 pKa = 3.58LYY271 pKa = 11.22SVPKK275 pKa = 7.78PTEE278 pKa = 3.71IEE280 pKa = 4.21TMSDD284 pKa = 2.71HH285 pKa = 7.0LNNHH289 pKa = 6.38KK290 pKa = 10.93SGDD293 pKa = 3.65TFPYY297 pKa = 8.9VSVVGTEE304 pKa = 4.75DD305 pKa = 3.14EE306 pKa = 4.6TKK308 pKa = 10.56DD309 pKa = 3.48GRR311 pKa = 11.84LSRR314 pKa = 11.84LRR316 pKa = 11.84AFRR319 pKa = 11.84SSYY322 pKa = 10.62GKK324 pKa = 9.92EE325 pKa = 3.99NKK327 pKa = 9.81PEE329 pKa = 3.66MRR331 pKa = 11.84INGVILDD338 pKa = 4.15EE339 pKa = 4.64LEE341 pKa = 4.11GTIKK345 pKa = 10.5VSNIGDD351 pKa = 3.77YY352 pKa = 10.99VNKK355 pKa = 10.42EE356 pKa = 4.05NLNKK360 pKa = 9.24QQRR363 pKa = 11.84SEE365 pKa = 4.1NKK367 pKa = 9.28RR368 pKa = 11.84FRR370 pKa = 11.84PLNEE374 pKa = 3.73ASFKK378 pKa = 10.89KK379 pKa = 10.11PYY381 pKa = 10.04LQIARR386 pKa = 11.84ITGEE390 pKa = 4.29YY391 pKa = 9.1VPLMSSTSDD400 pKa = 3.3YY401 pKa = 10.74TEE403 pKa = 5.1LYY405 pKa = 7.81FTLEE409 pKa = 4.43DD410 pKa = 4.84GRR412 pKa = 11.84LLDD415 pKa = 3.66NQVIIQSNKK424 pKa = 10.11LPTNQNGVFEE434 pKa = 5.05LSCDD438 pKa = 3.62YY439 pKa = 10.89CINLSDD445 pKa = 4.77INQLSLKK452 pKa = 9.72YY453 pKa = 10.04FLSRR457 pKa = 11.84PIMKK461 pKa = 10.3EE462 pKa = 3.73GFQWGAVSLTIRR474 pKa = 11.84VSEE477 pKa = 4.1SDD479 pKa = 3.52TPYY482 pKa = 9.9LTPKK486 pKa = 9.31VEE488 pKa = 3.77AMAIVRR494 pKa = 11.84IPFSTLEE501 pKa = 4.14EE502 pKa = 3.99QSKK505 pKa = 11.23DD506 pKa = 3.34PDD508 pKa = 3.58HH509 pKa = 7.08ADD511 pKa = 3.15VVFTAKK517 pKa = 10.21QVDD520 pKa = 3.93KK521 pKa = 11.18FKK523 pKa = 10.87EE524 pKa = 4.07LYY526 pKa = 10.02RR527 pKa = 11.84AGDD530 pKa = 3.42VMDD533 pKa = 4.0IDD535 pKa = 3.63EE536 pKa = 4.94AKK538 pKa = 10.42RR539 pKa = 11.84EE540 pKa = 3.78RR541 pKa = 11.84STINSYY547 pKa = 11.0SKK549 pKa = 9.02STIRR553 pKa = 11.84GVVKK557 pKa = 10.81GEE559 pKa = 4.24AGPSHH564 pKa = 7.42LGNQSGWEE572 pKa = 3.99HH573 pKa = 6.49LKK575 pKa = 11.18GMVKK579 pKa = 10.02PRR581 pKa = 11.84VEE583 pKa = 4.19EE584 pKa = 4.44GQASISVASGSEE596 pKa = 4.0EE597 pKa = 3.72VDD599 pKa = 2.56IDD601 pKa = 3.98VPSTTRR607 pKa = 11.84EE608 pKa = 3.98EE609 pKa = 4.39YY610 pKa = 10.39EE611 pKa = 4.06KK612 pKa = 10.6QQEE615 pKa = 4.25LLRR618 pKa = 11.84QQFQQTFSDD627 pKa = 3.72TTEE630 pKa = 4.28EE631 pKa = 4.15IPRR634 pKa = 11.84SEE636 pKa = 4.69LNRR639 pKa = 11.84SSSPINSDD647 pKa = 3.28PLSSVEE653 pKa = 3.97EE654 pKa = 4.2SSPPRR659 pKa = 11.84SPIKK663 pKa = 10.53SAMKK667 pKa = 9.81KK668 pKa = 9.79PRR670 pKa = 11.84FIEE673 pKa = 4.32DD674 pKa = 3.58TLKK677 pKa = 11.08KK678 pKa = 10.01PDD680 pKa = 3.66TSDD683 pKa = 3.34VYY685 pKa = 11.27QFNN688 pKa = 3.79
Molecular weight: 78.33 kDa
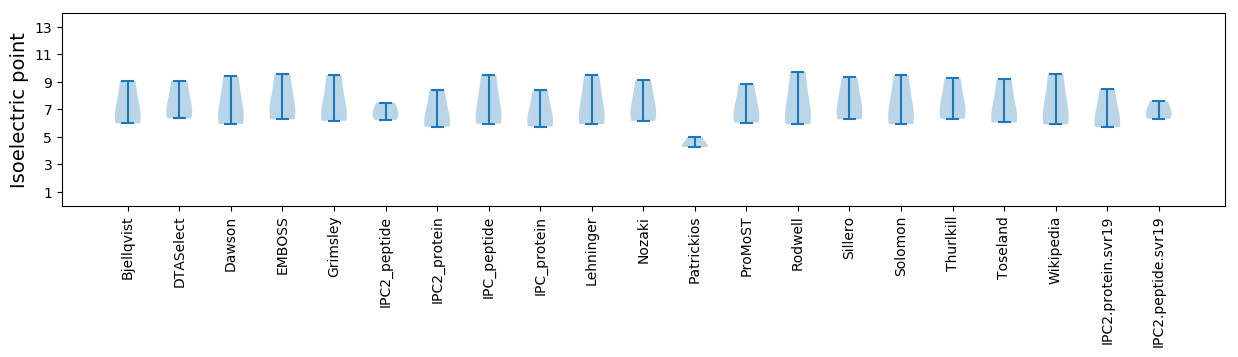
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A223PR01|A0A223PR01_9VIRU Replicase OS=Laurel Lake virus OX=2027354 PE=4 SV=1
MM1 pKa = 7.28TSALSVYY8 pKa = 9.78TMISAADD15 pKa = 3.58TTGVEE20 pKa = 4.25EE21 pKa = 4.65FSTLFAYY28 pKa = 10.03EE29 pKa = 4.11GFNPEE34 pKa = 4.44MIHH37 pKa = 6.02SHH39 pKa = 5.69FAKK42 pKa = 10.39IMTEE46 pKa = 3.7KK47 pKa = 10.87GIGEE51 pKa = 4.2MEE53 pKa = 4.37FVNDD57 pKa = 3.36MRR59 pKa = 11.84ALITLGAMKK68 pKa = 10.63GNYY71 pKa = 7.09TMKK74 pKa = 10.46NAGKK78 pKa = 9.69ISEE81 pKa = 4.29AGRR84 pKa = 11.84TKK86 pKa = 11.15ADD88 pKa = 3.35GLYY91 pKa = 10.46KK92 pKa = 10.24KK93 pKa = 10.94YY94 pKa = 11.2NMKK97 pKa = 10.31QGSLGGDD104 pKa = 3.12KK105 pKa = 10.56KK106 pKa = 11.08AVILPRR112 pKa = 11.84VLSAFPEE119 pKa = 4.34LTTKK123 pKa = 10.3VVLRR127 pKa = 11.84TPPRR131 pKa = 11.84DD132 pKa = 3.04FGTRR136 pKa = 11.84TTRR139 pKa = 11.84LPKK142 pKa = 10.13YY143 pKa = 9.25IKK145 pKa = 10.83NPVFPSLVPKK155 pKa = 9.43TLQGDD160 pKa = 3.9VAKK163 pKa = 9.97TFLWLYY169 pKa = 8.09TVYY172 pKa = 10.52SAEE175 pKa = 3.82QSLVISQEE183 pKa = 3.59KK184 pKa = 10.42DD185 pKa = 3.28FNAAFTAQKK194 pKa = 10.47QFVTIAFNSSVPDD207 pKa = 3.21EE208 pKa = 4.24TTRR211 pKa = 11.84IGMFKK216 pKa = 10.79AKK218 pKa = 10.7LEE220 pKa = 4.31DD221 pKa = 3.64LVQAISDD228 pKa = 3.89FKK230 pKa = 11.34DD231 pKa = 3.67FTDD234 pKa = 5.16GEE236 pKa = 4.84VPTNRR241 pKa = 11.84SDD243 pKa = 3.53ARR245 pKa = 11.84VARR248 pKa = 11.84DD249 pKa = 4.12AITTLL254 pKa = 3.5
MM1 pKa = 7.28TSALSVYY8 pKa = 9.78TMISAADD15 pKa = 3.58TTGVEE20 pKa = 4.25EE21 pKa = 4.65FSTLFAYY28 pKa = 10.03EE29 pKa = 4.11GFNPEE34 pKa = 4.44MIHH37 pKa = 6.02SHH39 pKa = 5.69FAKK42 pKa = 10.39IMTEE46 pKa = 3.7KK47 pKa = 10.87GIGEE51 pKa = 4.2MEE53 pKa = 4.37FVNDD57 pKa = 3.36MRR59 pKa = 11.84ALITLGAMKK68 pKa = 10.63GNYY71 pKa = 7.09TMKK74 pKa = 10.46NAGKK78 pKa = 9.69ISEE81 pKa = 4.29AGRR84 pKa = 11.84TKK86 pKa = 11.15ADD88 pKa = 3.35GLYY91 pKa = 10.46KK92 pKa = 10.24KK93 pKa = 10.94YY94 pKa = 11.2NMKK97 pKa = 10.31QGSLGGDD104 pKa = 3.12KK105 pKa = 10.56KK106 pKa = 11.08AVILPRR112 pKa = 11.84VLSAFPEE119 pKa = 4.34LTTKK123 pKa = 10.3VVLRR127 pKa = 11.84TPPRR131 pKa = 11.84DD132 pKa = 3.04FGTRR136 pKa = 11.84TTRR139 pKa = 11.84LPKK142 pKa = 10.13YY143 pKa = 9.25IKK145 pKa = 10.83NPVFPSLVPKK155 pKa = 9.43TLQGDD160 pKa = 3.9VAKK163 pKa = 9.97TFLWLYY169 pKa = 8.09TVYY172 pKa = 10.52SAEE175 pKa = 3.82QSLVISQEE183 pKa = 3.59KK184 pKa = 10.42DD185 pKa = 3.28FNAAFTAQKK194 pKa = 10.47QFVTIAFNSSVPDD207 pKa = 3.21EE208 pKa = 4.24TTRR211 pKa = 11.84IGMFKK216 pKa = 10.79AKK218 pKa = 10.7LEE220 pKa = 4.31DD221 pKa = 3.64LVQAISDD228 pKa = 3.89FKK230 pKa = 11.34DD231 pKa = 3.67FTDD234 pKa = 5.16GEE236 pKa = 4.84VPTNRR241 pKa = 11.84SDD243 pKa = 3.53ARR245 pKa = 11.84VARR248 pKa = 11.84DD249 pKa = 4.12AITTLL254 pKa = 3.5
Molecular weight: 28.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3326 |
254 |
2384 |
1108.7 |
126.54 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.788 ± 1.017 | 1.624 ± 0.485 |
6.133 ± 0.134 | 7.096 ± 0.426 |
4.63 ± 0.505 | 5.021 ± 0.247 |
2.195 ± 0.398 | 5.833 ± 0.328 |
7.817 ± 0.373 | 9.771 ± 0.656 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.277 ± 0.356 | 4.931 ± 0.347 |
3.578 ± 0.626 | 3.187 ± 0.39 |
4.901 ± 0.512 | 9.832 ± 0.816 |
6.133 ± 0.898 | 5.683 ± 0.316 |
0.992 ± 0.192 | 3.548 ± 0.077 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
