
Maize yellow striate virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Betarhabdovirinae; Cytorhabdovirus; Maize yellow striate cytorhabdovirus
Average proteome isoelectric point is 6.82
Get precalculated fractions of proteins
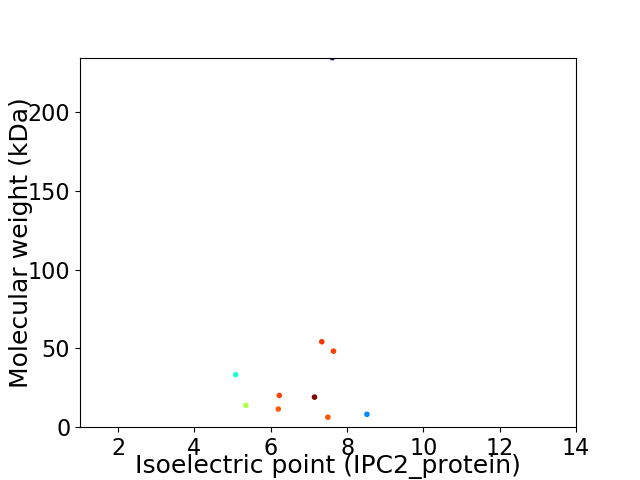
Virtual 2D-PAGE plot for 10 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2D1GTQ4|A0A2D1GTQ4_9RHAB N protein OS=Maize yellow striate virus OX=1168550 GN=N PE=4 SV=1
MM1 pKa = 7.65ASSSDD6 pKa = 3.51VQFLSGIPDD15 pKa = 4.94DD16 pKa = 3.75IHH18 pKa = 6.42PRR20 pKa = 11.84SYY22 pKa = 11.28DD23 pKa = 3.39LSADD27 pKa = 4.05GIFSNAEE34 pKa = 3.64EE35 pKa = 4.34EE36 pKa = 4.36EE37 pKa = 4.38VMEE40 pKa = 4.31EE41 pKa = 3.97RR42 pKa = 11.84QAEE45 pKa = 4.0QDD47 pKa = 3.52LPDD50 pKa = 4.5IEE52 pKa = 4.88EE53 pKa = 4.31EE54 pKa = 4.22EE55 pKa = 4.29PLVEE59 pKa = 5.18KK60 pKa = 9.77PAKK63 pKa = 10.07GEE65 pKa = 4.1KK66 pKa = 9.59FVSGTDD72 pKa = 3.45EE73 pKa = 4.18LVSILKK79 pKa = 10.31DD80 pKa = 3.3YY81 pKa = 11.34SNRR84 pKa = 11.84KK85 pKa = 8.06GMGMKK90 pKa = 10.13KK91 pKa = 9.4EE92 pKa = 4.06WVNIMKK98 pKa = 10.28RR99 pKa = 11.84RR100 pKa = 11.84FHH102 pKa = 6.65EE103 pKa = 4.15MGGKK107 pKa = 7.9MLKK110 pKa = 8.1THH112 pKa = 6.44VDD114 pKa = 4.21FFLLGIQAEE123 pKa = 4.55RR124 pKa = 11.84NMSVDD129 pKa = 3.9RR130 pKa = 11.84DD131 pKa = 4.06FKK133 pKa = 10.98DD134 pKa = 2.87TATRR138 pKa = 11.84LSDD141 pKa = 3.23EE142 pKa = 4.45VNRR145 pKa = 11.84VSGVGKK151 pKa = 10.61RR152 pKa = 11.84MLDD155 pKa = 3.23AQQKK159 pKa = 7.36MEE161 pKa = 4.68RR162 pKa = 11.84EE163 pKa = 4.3MEE165 pKa = 4.13QKK167 pKa = 9.57MKK169 pKa = 10.57EE170 pKa = 3.82ITAHH174 pKa = 5.34CKK176 pKa = 9.73KK177 pKa = 9.87MEE179 pKa = 4.01MMVAKK184 pKa = 10.77VEE186 pKa = 4.33TAVEE190 pKa = 4.08NASRR194 pKa = 11.84PSSISSWAMGGPSRR208 pKa = 11.84EE209 pKa = 4.09EE210 pKa = 3.86EE211 pKa = 4.04EE212 pKa = 4.0EE213 pKa = 3.83RR214 pKa = 11.84DD215 pKa = 3.48YY216 pKa = 11.87DD217 pKa = 4.48KK218 pKa = 10.87FLSMIGFEE226 pKa = 4.53DD227 pKa = 3.54KK228 pKa = 10.58HH229 pKa = 7.47IKK231 pKa = 8.91SATMKK236 pKa = 9.74KK237 pKa = 9.66CYY239 pKa = 9.24PAISDD244 pKa = 3.27EE245 pKa = 4.57MYY247 pKa = 9.56VHH249 pKa = 7.0VITGEE254 pKa = 3.88ADD256 pKa = 3.13HH257 pKa = 8.04ADD259 pKa = 3.16MTNYY263 pKa = 9.71YY264 pKa = 9.8RR265 pKa = 11.84LIMEE269 pKa = 4.22YY270 pKa = 10.94AEE272 pKa = 4.29SKK274 pKa = 9.9VLKK277 pKa = 9.21KK278 pKa = 10.69KK279 pKa = 10.58SVAASRR285 pKa = 11.84NDD287 pKa = 3.75PYY289 pKa = 11.06PGLL292 pKa = 3.99
MM1 pKa = 7.65ASSSDD6 pKa = 3.51VQFLSGIPDD15 pKa = 4.94DD16 pKa = 3.75IHH18 pKa = 6.42PRR20 pKa = 11.84SYY22 pKa = 11.28DD23 pKa = 3.39LSADD27 pKa = 4.05GIFSNAEE34 pKa = 3.64EE35 pKa = 4.34EE36 pKa = 4.36EE37 pKa = 4.38VMEE40 pKa = 4.31EE41 pKa = 3.97RR42 pKa = 11.84QAEE45 pKa = 4.0QDD47 pKa = 3.52LPDD50 pKa = 4.5IEE52 pKa = 4.88EE53 pKa = 4.31EE54 pKa = 4.22EE55 pKa = 4.29PLVEE59 pKa = 5.18KK60 pKa = 9.77PAKK63 pKa = 10.07GEE65 pKa = 4.1KK66 pKa = 9.59FVSGTDD72 pKa = 3.45EE73 pKa = 4.18LVSILKK79 pKa = 10.31DD80 pKa = 3.3YY81 pKa = 11.34SNRR84 pKa = 11.84KK85 pKa = 8.06GMGMKK90 pKa = 10.13KK91 pKa = 9.4EE92 pKa = 4.06WVNIMKK98 pKa = 10.28RR99 pKa = 11.84RR100 pKa = 11.84FHH102 pKa = 6.65EE103 pKa = 4.15MGGKK107 pKa = 7.9MLKK110 pKa = 8.1THH112 pKa = 6.44VDD114 pKa = 4.21FFLLGIQAEE123 pKa = 4.55RR124 pKa = 11.84NMSVDD129 pKa = 3.9RR130 pKa = 11.84DD131 pKa = 4.06FKK133 pKa = 10.98DD134 pKa = 2.87TATRR138 pKa = 11.84LSDD141 pKa = 3.23EE142 pKa = 4.45VNRR145 pKa = 11.84VSGVGKK151 pKa = 10.61RR152 pKa = 11.84MLDD155 pKa = 3.23AQQKK159 pKa = 7.36MEE161 pKa = 4.68RR162 pKa = 11.84EE163 pKa = 4.3MEE165 pKa = 4.13QKK167 pKa = 9.57MKK169 pKa = 10.57EE170 pKa = 3.82ITAHH174 pKa = 5.34CKK176 pKa = 9.73KK177 pKa = 9.87MEE179 pKa = 4.01MMVAKK184 pKa = 10.77VEE186 pKa = 4.33TAVEE190 pKa = 4.08NASRR194 pKa = 11.84PSSISSWAMGGPSRR208 pKa = 11.84EE209 pKa = 4.09EE210 pKa = 3.86EE211 pKa = 4.04EE212 pKa = 4.0EE213 pKa = 3.83RR214 pKa = 11.84DD215 pKa = 3.48YY216 pKa = 11.87DD217 pKa = 4.48KK218 pKa = 10.87FLSMIGFEE226 pKa = 4.53DD227 pKa = 3.54KK228 pKa = 10.58HH229 pKa = 7.47IKK231 pKa = 8.91SATMKK236 pKa = 9.74KK237 pKa = 9.66CYY239 pKa = 9.24PAISDD244 pKa = 3.27EE245 pKa = 4.57MYY247 pKa = 9.56VHH249 pKa = 7.0VITGEE254 pKa = 3.88ADD256 pKa = 3.13HH257 pKa = 8.04ADD259 pKa = 3.16MTNYY263 pKa = 9.71YY264 pKa = 9.8RR265 pKa = 11.84LIMEE269 pKa = 4.22YY270 pKa = 10.94AEE272 pKa = 4.29SKK274 pKa = 9.9VLKK277 pKa = 9.21KK278 pKa = 10.69KK279 pKa = 10.58SVAASRR285 pKa = 11.84NDD287 pKa = 3.75PYY289 pKa = 11.06PGLL292 pKa = 3.99
Molecular weight: 33.38 kDa
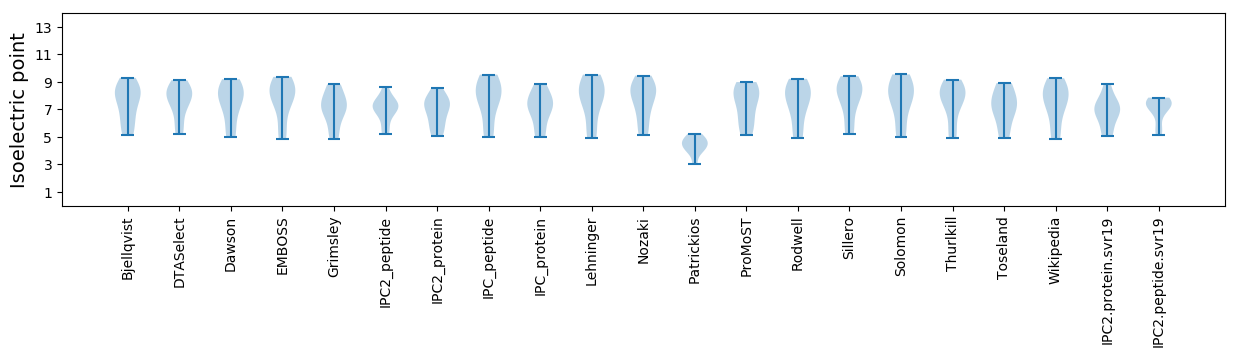
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2D1GTP8|A0A2D1GTP8_9RHAB P protein OS=Maize yellow striate virus OX=1168550 GN=P PE=4 SV=1
MM1 pKa = 7.7WNFLLHH7 pKa = 5.32VLRR10 pKa = 11.84DD11 pKa = 3.62GSDD14 pKa = 3.36RR15 pKa = 11.84TNMSSSWTVVSLSAYY30 pKa = 9.72AILVFIISISICRR43 pKa = 11.84LKK45 pKa = 10.79VYY47 pKa = 7.94QLRR50 pKa = 11.84LIVIISLCHH59 pKa = 5.6LVSAIGSAFKK69 pKa = 10.5EE70 pKa = 4.34WTT72 pKa = 3.41
MM1 pKa = 7.7WNFLLHH7 pKa = 5.32VLRR10 pKa = 11.84DD11 pKa = 3.62GSDD14 pKa = 3.36RR15 pKa = 11.84TNMSSSWTVVSLSAYY30 pKa = 9.72AILVFIISISICRR43 pKa = 11.84LKK45 pKa = 10.79VYY47 pKa = 7.94QLRR50 pKa = 11.84LIVIISLCHH59 pKa = 5.6LVSAIGSAFKK69 pKa = 10.5EE70 pKa = 4.34WTT72 pKa = 3.41
Molecular weight: 8.19 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3954 |
51 |
2056 |
395.4 |
44.99 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.982 ± 0.645 | 1.669 ± 0.477 |
5.387 ± 0.42 | 6.5 ± 0.703 |
3.92 ± 0.249 | 5.589 ± 0.359 |
2.15 ± 0.313 | 7.031 ± 0.49 |
6.651 ± 0.529 | 9.611 ± 1.026 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.288 ± 0.497 | 4.401 ± 0.367 |
4.299 ± 0.44 | 2.731 ± 0.314 |
5.438 ± 0.327 | 8.574 ± 0.596 |
5.918 ± 0.694 | 6.348 ± 0.451 |
1.593 ± 0.285 | 3.92 ± 0.422 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
