
Desulfarculus baarsii (strain ATCC 33931 / DSM 2075 / LMG 7858 / VKM B-1802 / 2st14)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Desulfarculales; Desulfarculaceae; Desulfarculus; Desulfarculus baarsii
Average proteome isoelectric point is 6.66
Get precalculated fractions of proteins
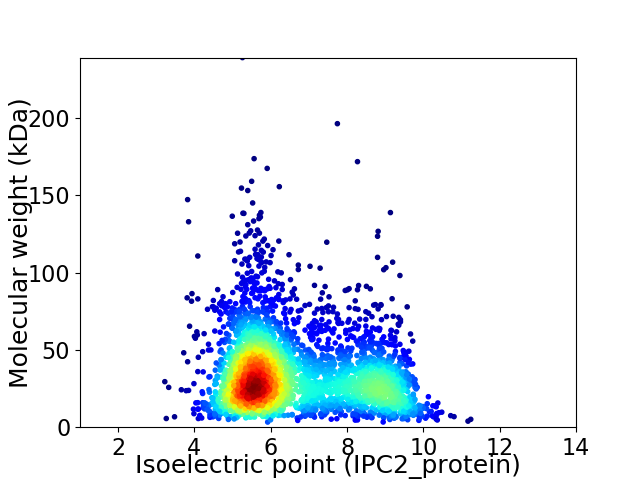
Virtual 2D-PAGE plot for 3268 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E1QHJ2|E1QHJ2_DESB2 Binding-protein-dependent transport systems inner membrane component OS=Desulfarculus baarsii (strain ATCC 33931 / DSM 2075 / LMG 7858 / VKM B-1802 / 2st14) OX=644282 GN=Deba_1667 PE=3 SV=1
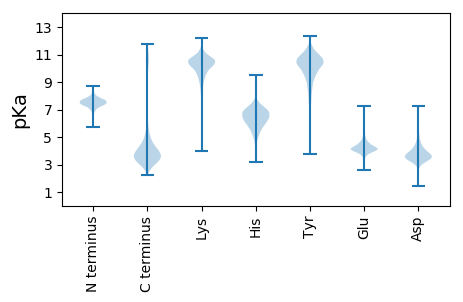
MM1 pKa = 7.1QATSFFNGVSGLKK14 pKa = 10.4SFSQGLNIVADD25 pKa = 3.71NLANSNTYY33 pKa = 10.0GYY35 pKa = 10.09KK36 pKa = 9.78SSRR39 pKa = 11.84AEE41 pKa = 3.95FADD44 pKa = 3.21VLYY47 pKa = 10.92RR48 pKa = 11.84EE49 pKa = 4.49LSYY52 pKa = 10.79TGSDD56 pKa = 3.14LTTEE60 pKa = 4.34IDD62 pKa = 3.68QVGQGATVWSHH73 pKa = 5.67QLMNQGAIQEE83 pKa = 4.4TGRR86 pKa = 11.84TLDD89 pKa = 4.28LAIDD93 pKa = 3.84GNGFFTVKK101 pKa = 10.64NLDD104 pKa = 3.71TEE106 pKa = 4.16EE107 pKa = 4.65LYY109 pKa = 9.3YY110 pKa = 10.78TRR112 pKa = 11.84AGQFGVDD119 pKa = 3.25GVVGQEE125 pKa = 3.74GFIINDD131 pKa = 2.86QGYY134 pKa = 8.69RR135 pKa = 11.84LQGFAIGDD143 pKa = 3.77DD144 pKa = 3.95GEE146 pKa = 4.85PIVGNLIDD154 pKa = 4.06LQIPVEE160 pKa = 4.11NLPGEE165 pKa = 4.18ATTIVGLGVNLNPADD180 pKa = 3.63TRR182 pKa = 11.84VNQVATDD189 pKa = 3.78IDD191 pKa = 4.19PEE193 pKa = 4.23VSGTYY198 pKa = 9.88NYY200 pKa = 10.81SSSTTVYY207 pKa = 10.37DD208 pKa = 4.03ANGDD212 pKa = 3.56THH214 pKa = 6.93QISVYY219 pKa = 7.03YY220 pKa = 10.23QRR222 pKa = 11.84VDD224 pKa = 3.81DD225 pKa = 4.2YY226 pKa = 11.73AGTVPEE232 pKa = 5.02GGQTVWKK239 pKa = 10.09ASTFEE244 pKa = 4.26TQDD247 pKa = 3.29GEE249 pKa = 4.69QVANPADD256 pKa = 3.9PANTFYY262 pKa = 11.5LHH264 pKa = 5.95FTDD267 pKa = 3.83TGALAGVTDD276 pKa = 3.85STGATVSADD285 pKa = 3.97SIGLTMDD292 pKa = 4.01FGEE295 pKa = 5.26AGQQAITLDD304 pKa = 3.98FAPAAGQATTQVAEE318 pKa = 5.08GYY320 pKa = 7.76STSTNTQDD328 pKa = 3.38GFAEE332 pKa = 4.84GGLEE336 pKa = 4.0SVAVSEE342 pKa = 4.96DD343 pKa = 3.73GFVTAYY349 pKa = 9.92YY350 pKa = 11.0SNGEE354 pKa = 4.12MVDD357 pKa = 3.48VGVVALTTFASPGNLRR373 pKa = 11.84RR374 pKa = 11.84EE375 pKa = 4.16GDD377 pKa = 3.72NLWAYY382 pKa = 10.44DD383 pKa = 3.8ADD385 pKa = 4.43AGEE388 pKa = 4.26IWVGQPTDD396 pKa = 3.36EE397 pKa = 4.28EE398 pKa = 4.51FAMGAIEE405 pKa = 4.41DD406 pKa = 3.95QSLEE410 pKa = 4.08TSTVDD415 pKa = 3.36TATEE419 pKa = 3.92MMNMIIYY426 pKa = 9.75QRR428 pKa = 11.84AFQASSKK435 pKa = 9.68TVTTSDD441 pKa = 3.47EE442 pKa = 4.31MIKK445 pKa = 9.14TAINMKK451 pKa = 8.72TT452 pKa = 3.05
MM1 pKa = 7.1QATSFFNGVSGLKK14 pKa = 10.4SFSQGLNIVADD25 pKa = 3.71NLANSNTYY33 pKa = 10.0GYY35 pKa = 10.09KK36 pKa = 9.78SSRR39 pKa = 11.84AEE41 pKa = 3.95FADD44 pKa = 3.21VLYY47 pKa = 10.92RR48 pKa = 11.84EE49 pKa = 4.49LSYY52 pKa = 10.79TGSDD56 pKa = 3.14LTTEE60 pKa = 4.34IDD62 pKa = 3.68QVGQGATVWSHH73 pKa = 5.67QLMNQGAIQEE83 pKa = 4.4TGRR86 pKa = 11.84TLDD89 pKa = 4.28LAIDD93 pKa = 3.84GNGFFTVKK101 pKa = 10.64NLDD104 pKa = 3.71TEE106 pKa = 4.16EE107 pKa = 4.65LYY109 pKa = 9.3YY110 pKa = 10.78TRR112 pKa = 11.84AGQFGVDD119 pKa = 3.25GVVGQEE125 pKa = 3.74GFIINDD131 pKa = 2.86QGYY134 pKa = 8.69RR135 pKa = 11.84LQGFAIGDD143 pKa = 3.77DD144 pKa = 3.95GEE146 pKa = 4.85PIVGNLIDD154 pKa = 4.06LQIPVEE160 pKa = 4.11NLPGEE165 pKa = 4.18ATTIVGLGVNLNPADD180 pKa = 3.63TRR182 pKa = 11.84VNQVATDD189 pKa = 3.78IDD191 pKa = 4.19PEE193 pKa = 4.23VSGTYY198 pKa = 9.88NYY200 pKa = 10.81SSSTTVYY207 pKa = 10.37DD208 pKa = 4.03ANGDD212 pKa = 3.56THH214 pKa = 6.93QISVYY219 pKa = 7.03YY220 pKa = 10.23QRR222 pKa = 11.84VDD224 pKa = 3.81DD225 pKa = 4.2YY226 pKa = 11.73AGTVPEE232 pKa = 5.02GGQTVWKK239 pKa = 10.09ASTFEE244 pKa = 4.26TQDD247 pKa = 3.29GEE249 pKa = 4.69QVANPADD256 pKa = 3.9PANTFYY262 pKa = 11.5LHH264 pKa = 5.95FTDD267 pKa = 3.83TGALAGVTDD276 pKa = 3.85STGATVSADD285 pKa = 3.97SIGLTMDD292 pKa = 4.01FGEE295 pKa = 5.26AGQQAITLDD304 pKa = 3.98FAPAAGQATTQVAEE318 pKa = 5.08GYY320 pKa = 7.76STSTNTQDD328 pKa = 3.38GFAEE332 pKa = 4.84GGLEE336 pKa = 4.0SVAVSEE342 pKa = 4.96DD343 pKa = 3.73GFVTAYY349 pKa = 9.92YY350 pKa = 11.0SNGEE354 pKa = 4.12MVDD357 pKa = 3.48VGVVALTTFASPGNLRR373 pKa = 11.84RR374 pKa = 11.84EE375 pKa = 4.16GDD377 pKa = 3.72NLWAYY382 pKa = 10.44DD383 pKa = 3.8ADD385 pKa = 4.43AGEE388 pKa = 4.26IWVGQPTDD396 pKa = 3.36EE397 pKa = 4.28EE398 pKa = 4.51FAMGAIEE405 pKa = 4.41DD406 pKa = 3.95QSLEE410 pKa = 4.08TSTVDD415 pKa = 3.36TATEE419 pKa = 3.92MMNMIIYY426 pKa = 9.75QRR428 pKa = 11.84AFQASSKK435 pKa = 9.68TVTTSDD441 pKa = 3.47EE442 pKa = 4.31MIKK445 pKa = 9.14TAINMKK451 pKa = 8.72TT452 pKa = 3.05
Molecular weight: 48.14 kDa
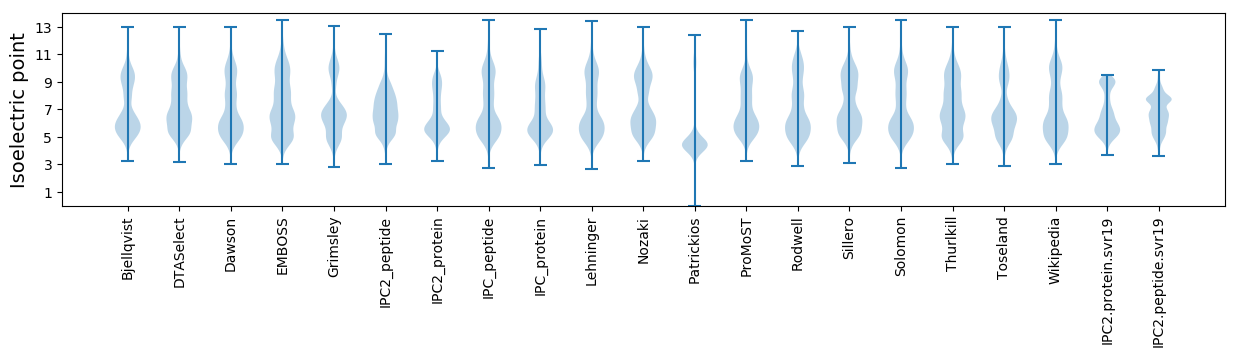
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E1QGF0|E1QGF0_DESB2 ATP synthase subunit alpha OS=Desulfarculus baarsii (strain ATCC 33931 / DSM 2075 / LMG 7858 / VKM B-1802 / 2st14) OX=644282 GN=atpA PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.2RR12 pKa = 11.84KK13 pKa = 7.55RR14 pKa = 11.84THH16 pKa = 5.91GFLVRR21 pKa = 11.84SRR23 pKa = 11.84SKK25 pKa = 10.78AGRR28 pKa = 11.84AVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.25GRR39 pKa = 11.84ARR41 pKa = 11.84LSVV44 pKa = 3.12
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.2RR12 pKa = 11.84KK13 pKa = 7.55RR14 pKa = 11.84THH16 pKa = 5.91GFLVRR21 pKa = 11.84SRR23 pKa = 11.84SKK25 pKa = 10.78AGRR28 pKa = 11.84AVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.25GRR39 pKa = 11.84ARR41 pKa = 11.84LSVV44 pKa = 3.12
Molecular weight: 5.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1096967 |
30 |
2142 |
335.7 |
36.5 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.682 ± 0.07 | 1.27 ± 0.017 |
5.504 ± 0.031 | 5.793 ± 0.035 |
3.402 ± 0.023 | 8.842 ± 0.043 |
2.001 ± 0.018 | 4.452 ± 0.031 |
3.666 ± 0.036 | 11.245 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.667 ± 0.019 | 2.538 ± 0.026 |
5.178 ± 0.032 | 4.094 ± 0.027 |
7.094 ± 0.036 | 4.588 ± 0.029 |
4.124 ± 0.026 | 7.261 ± 0.037 |
1.318 ± 0.019 | 2.281 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
