
Shuangao sobemo-like virus 1
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
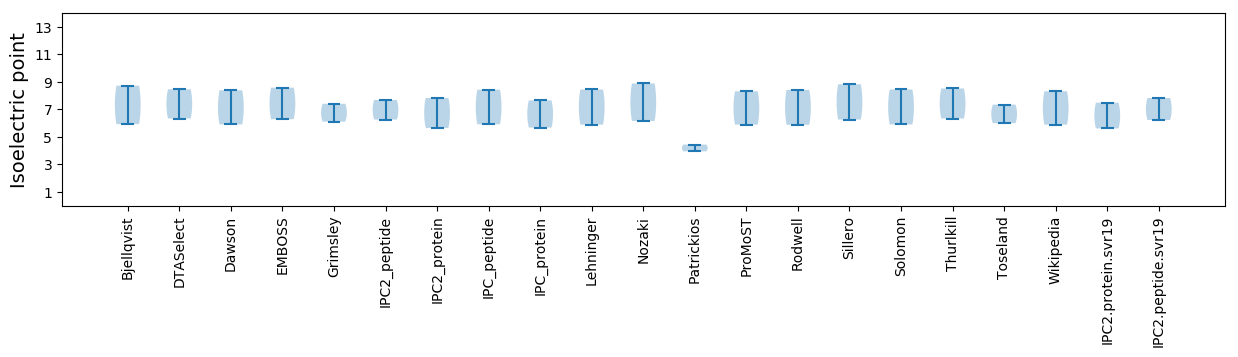
Average proteome isoelectric point is 6.55
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A1L3KEE6|A0A1L3KEE6_9VIRU RNA-directed RNA polymerase OS=Shuangao sobemo-like virus 1 OX=1923474 PE=4 SV=1
MM1 pKa = 7.67RR2 pKa = 11.84SIAAHH7 pKa = 5.73AGLRR11 pKa = 11.84EE12 pKa = 4.06GLSDD16 pKa = 4.32GPCNSIRR23 pKa = 11.84TKK25 pKa = 10.23MIQTVIEE32 pKa = 4.28RR33 pKa = 11.84NMAAQWEE40 pKa = 4.42IPPDD44 pKa = 3.71FLEE47 pKa = 4.36RR48 pKa = 11.84THH50 pKa = 7.0FDD52 pKa = 3.02RR53 pKa = 11.84VVAAIDD59 pKa = 3.51MTSTPGYY66 pKa = 9.3PYY68 pKa = 10.12IRR70 pKa = 11.84SATTNRR76 pKa = 11.84QYY78 pKa = 11.53FNADD82 pKa = 3.32DD83 pKa = 3.76PVKK86 pKa = 10.61FRR88 pKa = 11.84EE89 pKa = 4.29TCDD92 pKa = 3.79AIYY95 pKa = 8.46EE96 pKa = 4.49TVCAKK101 pKa = 10.09IDD103 pKa = 3.39GRR105 pKa = 11.84MGADD109 pKa = 4.18HH110 pKa = 7.0IRR112 pKa = 11.84LFIKK116 pKa = 10.69GEE118 pKa = 3.98PLPQHH123 pKa = 6.1KK124 pKa = 10.59VEE126 pKa = 4.19EE127 pKa = 4.47GRR129 pKa = 11.84LRR131 pKa = 11.84LISSVSVVDD140 pKa = 4.17QIVDD144 pKa = 3.41AMLHH148 pKa = 5.66GACNEE153 pKa = 3.84ALKK156 pKa = 10.49EE157 pKa = 3.68KK158 pKa = 10.16HH159 pKa = 6.67AYY161 pKa = 7.01VTPKK165 pKa = 10.62VGWSPYY171 pKa = 8.98GGGWKK176 pKa = 9.94IIPKK180 pKa = 8.49EE181 pKa = 3.52GWMALDD187 pKa = 3.67KK188 pKa = 11.03KK189 pKa = 10.79AWDD192 pKa = 3.51WSVKK196 pKa = 9.55PWILEE201 pKa = 3.81AEE203 pKa = 4.16LEE205 pKa = 4.1IRR207 pKa = 11.84IGLCTNINEE216 pKa = 4.31KK217 pKa = 9.84WLTLARR223 pKa = 11.84KK224 pKa = 9.48RR225 pKa = 11.84YY226 pKa = 7.67EE227 pKa = 3.8QLFVEE232 pKa = 5.43PIFVTSGGLLLQQIEE247 pKa = 4.21GGIIKK252 pKa = 10.31SGMVCTITTNCFGQDD267 pKa = 2.83ITHH270 pKa = 5.77VTTCYY275 pKa = 10.59TNGLLADD282 pKa = 4.18WLYY285 pKa = 12.13SMGDD289 pKa = 3.62DD290 pKa = 3.55TLQPPLEE297 pKa = 4.32EE298 pKa = 4.39PEE300 pKa = 3.88RR301 pKa = 11.84TEE303 pKa = 3.93YY304 pKa = 11.33LEE306 pKa = 4.23TLSQYY311 pKa = 10.48CHH313 pKa = 6.82IKK315 pKa = 10.34PGVGTNEE322 pKa = 3.85FAGFRR327 pKa = 11.84YY328 pKa = 10.06GRR330 pKa = 11.84GIEE333 pKa = 3.82PLYY336 pKa = 10.33RR337 pKa = 11.84PRR339 pKa = 11.84HH340 pKa = 5.59AYY342 pKa = 9.41QLLHH346 pKa = 6.75FDD348 pKa = 4.66DD349 pKa = 5.21RR350 pKa = 11.84YY351 pKa = 10.86KK352 pKa = 10.86EE353 pKa = 3.9QLAASYY359 pKa = 9.29TLLYY363 pKa = 10.1HH364 pKa = 7.61RR365 pKa = 11.84SSCRR369 pKa = 11.84NWMEE373 pKa = 4.27KK374 pKa = 10.46FFTEE378 pKa = 5.14LGISFPARR386 pKa = 11.84NIRR389 pKa = 11.84DD390 pKa = 3.94IIWDD394 pKa = 3.65SEE396 pKa = 4.21EE397 pKa = 3.76
MM1 pKa = 7.67RR2 pKa = 11.84SIAAHH7 pKa = 5.73AGLRR11 pKa = 11.84EE12 pKa = 4.06GLSDD16 pKa = 4.32GPCNSIRR23 pKa = 11.84TKK25 pKa = 10.23MIQTVIEE32 pKa = 4.28RR33 pKa = 11.84NMAAQWEE40 pKa = 4.42IPPDD44 pKa = 3.71FLEE47 pKa = 4.36RR48 pKa = 11.84THH50 pKa = 7.0FDD52 pKa = 3.02RR53 pKa = 11.84VVAAIDD59 pKa = 3.51MTSTPGYY66 pKa = 9.3PYY68 pKa = 10.12IRR70 pKa = 11.84SATTNRR76 pKa = 11.84QYY78 pKa = 11.53FNADD82 pKa = 3.32DD83 pKa = 3.76PVKK86 pKa = 10.61FRR88 pKa = 11.84EE89 pKa = 4.29TCDD92 pKa = 3.79AIYY95 pKa = 8.46EE96 pKa = 4.49TVCAKK101 pKa = 10.09IDD103 pKa = 3.39GRR105 pKa = 11.84MGADD109 pKa = 4.18HH110 pKa = 7.0IRR112 pKa = 11.84LFIKK116 pKa = 10.69GEE118 pKa = 3.98PLPQHH123 pKa = 6.1KK124 pKa = 10.59VEE126 pKa = 4.19EE127 pKa = 4.47GRR129 pKa = 11.84LRR131 pKa = 11.84LISSVSVVDD140 pKa = 4.17QIVDD144 pKa = 3.41AMLHH148 pKa = 5.66GACNEE153 pKa = 3.84ALKK156 pKa = 10.49EE157 pKa = 3.68KK158 pKa = 10.16HH159 pKa = 6.67AYY161 pKa = 7.01VTPKK165 pKa = 10.62VGWSPYY171 pKa = 8.98GGGWKK176 pKa = 9.94IIPKK180 pKa = 8.49EE181 pKa = 3.52GWMALDD187 pKa = 3.67KK188 pKa = 11.03KK189 pKa = 10.79AWDD192 pKa = 3.51WSVKK196 pKa = 9.55PWILEE201 pKa = 3.81AEE203 pKa = 4.16LEE205 pKa = 4.1IRR207 pKa = 11.84IGLCTNINEE216 pKa = 4.31KK217 pKa = 9.84WLTLARR223 pKa = 11.84KK224 pKa = 9.48RR225 pKa = 11.84YY226 pKa = 7.67EE227 pKa = 3.8QLFVEE232 pKa = 5.43PIFVTSGGLLLQQIEE247 pKa = 4.21GGIIKK252 pKa = 10.31SGMVCTITTNCFGQDD267 pKa = 2.83ITHH270 pKa = 5.77VTTCYY275 pKa = 10.59TNGLLADD282 pKa = 4.18WLYY285 pKa = 12.13SMGDD289 pKa = 3.62DD290 pKa = 3.55TLQPPLEE297 pKa = 4.32EE298 pKa = 4.39PEE300 pKa = 3.88RR301 pKa = 11.84TEE303 pKa = 3.93YY304 pKa = 11.33LEE306 pKa = 4.23TLSQYY311 pKa = 10.48CHH313 pKa = 6.82IKK315 pKa = 10.34PGVGTNEE322 pKa = 3.85FAGFRR327 pKa = 11.84YY328 pKa = 10.06GRR330 pKa = 11.84GIEE333 pKa = 3.82PLYY336 pKa = 10.33RR337 pKa = 11.84PRR339 pKa = 11.84HH340 pKa = 5.59AYY342 pKa = 9.41QLLHH346 pKa = 6.75FDD348 pKa = 4.66DD349 pKa = 5.21RR350 pKa = 11.84YY351 pKa = 10.86KK352 pKa = 10.86EE353 pKa = 3.9QLAASYY359 pKa = 9.29TLLYY363 pKa = 10.1HH364 pKa = 7.61RR365 pKa = 11.84SSCRR369 pKa = 11.84NWMEE373 pKa = 4.27KK374 pKa = 10.46FFTEE378 pKa = 5.14LGISFPARR386 pKa = 11.84NIRR389 pKa = 11.84DD390 pKa = 3.94IIWDD394 pKa = 3.65SEE396 pKa = 4.21EE397 pKa = 3.76
Molecular weight: 45.46 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KEE6|A0A1L3KEE6_9VIRU RNA-directed RNA polymerase OS=Shuangao sobemo-like virus 1 OX=1923474 PE=4 SV=1
MM1 pKa = 7.66LNALWNQRR9 pKa = 11.84KK10 pKa = 9.09LFVVGITLYY19 pKa = 10.9LVLSGWSGLPLHH31 pKa = 6.04TLVFRR36 pKa = 11.84TVRR39 pKa = 11.84YY40 pKa = 8.85LVWKK44 pKa = 9.9YY45 pKa = 9.02VLRR48 pKa = 11.84DD49 pKa = 4.05TIAGQFLEE57 pKa = 4.43EE58 pKa = 4.58LSSEE62 pKa = 4.13ILGKK66 pKa = 10.27EE67 pKa = 4.01MPPPPPPPPPKK78 pKa = 9.96PIPPLTDD85 pKa = 2.82RR86 pKa = 11.84LYY88 pKa = 10.88EE89 pKa = 4.04DD90 pKa = 3.97WIGLYY95 pKa = 10.55GRR97 pKa = 11.84TVNSTTRR104 pKa = 11.84VLEE107 pKa = 3.8KK108 pKa = 10.28AYY110 pKa = 10.48YY111 pKa = 10.21SSWTSWFHH119 pKa = 5.09MACNTIAIIVAIRR132 pKa = 11.84TLFVAISRR140 pKa = 11.84IIPGSPRR147 pKa = 11.84TAVATWAKK155 pKa = 10.01CKK157 pKa = 10.04IRR159 pKa = 11.84GIHH162 pKa = 5.8YY163 pKa = 7.45EE164 pKa = 3.77AAIQGSLLRR173 pKa = 11.84KK174 pKa = 8.41ATIPEE179 pKa = 3.99WQVEE183 pKa = 4.3VLTPGFLSDD192 pKa = 3.23THH194 pKa = 7.11VGWGVRR200 pKa = 11.84VVDD203 pKa = 3.71TLVVPTHH210 pKa = 5.88VLVEE214 pKa = 4.43AGPNYY219 pKa = 10.21LLKK222 pKa = 10.91GRR224 pKa = 11.84TGSVLCNGLPRR235 pKa = 11.84DD236 pKa = 3.81SRR238 pKa = 11.84IMQDD242 pKa = 2.42ISYY245 pKa = 10.39VPVEE249 pKa = 3.82VSTWSKK255 pKa = 11.05LGTPSVKK262 pKa = 9.9RR263 pKa = 11.84KK264 pKa = 10.08RR265 pKa = 11.84IDD267 pKa = 3.25VEE269 pKa = 4.51TVQSVSCCGLEE280 pKa = 4.33GASTGPLRR288 pKa = 11.84QTRR291 pKa = 11.84VRR293 pKa = 11.84GLMYY297 pKa = 10.83YY298 pKa = 10.1EE299 pKa = 4.59GTTMPGMSGAGYY311 pKa = 10.51YY312 pKa = 9.7IGNTCLGIHH321 pKa = 6.85LGASTNVNCGATIMVPFAEE340 pKa = 4.7IPAPNNLVVQVEE352 pKa = 4.81RR353 pKa = 11.84SAKK356 pKa = 9.56RR357 pKa = 11.84PKK359 pKa = 9.1KK360 pKa = 8.51TRR362 pKa = 11.84PPSGTSADD370 pKa = 3.89PTYY373 pKa = 11.02HH374 pKa = 7.78AMDD377 pKa = 3.57NTKK380 pKa = 8.8TAWGYY385 pKa = 11.02ADD387 pKa = 3.68IRR389 pKa = 11.84EE390 pKa = 4.4ALDD393 pKa = 4.1DD394 pKa = 3.89YY395 pKa = 11.73DD396 pKa = 5.56RR397 pKa = 11.84FIGPSDD403 pKa = 3.48WNNPTDD409 pKa = 5.34DD410 pKa = 4.12FDD412 pKa = 5.13YY413 pKa = 11.27DD414 pKa = 3.17QDD416 pKa = 3.59LGYY419 pKa = 10.6EE420 pKa = 4.23YY421 pKa = 10.62EE422 pKa = 4.3EE423 pKa = 4.27EE424 pKa = 3.92AATRR428 pKa = 11.84GNTLMEE434 pKa = 4.49QIRR437 pKa = 11.84RR438 pKa = 11.84MSVEE442 pKa = 4.26DD443 pKa = 3.28RR444 pKa = 11.84KK445 pKa = 11.09LWMDD449 pKa = 4.6SINAVATEE457 pKa = 4.0EE458 pKa = 4.06AVAARR463 pKa = 11.84STPVANPLPSVRR475 pKa = 11.84PMVQTRR481 pKa = 11.84QGVPRR486 pKa = 11.84QVTFDD491 pKa = 3.86EE492 pKa = 5.24IIVHH496 pKa = 5.62NQTGVGTSTTIHH508 pKa = 6.0TQPQPQRR515 pKa = 11.84LAGPPHH521 pKa = 7.12LGDD524 pKa = 4.74LVRR527 pKa = 11.84DD528 pKa = 4.22LIKK531 pKa = 10.51RR532 pKa = 11.84VTILEE537 pKa = 3.86QKK539 pKa = 9.76MIQRR543 pKa = 11.84DD544 pKa = 3.42EE545 pKa = 4.8RR546 pKa = 11.84MVEE549 pKa = 3.6IEE551 pKa = 4.15NAVGTLQDD559 pKa = 3.45AVAKK563 pKa = 10.01YY564 pKa = 9.93GSPEE568 pKa = 3.84EE569 pKa = 4.16LEE571 pKa = 4.15KK572 pKa = 11.12LRR574 pKa = 11.84NDD576 pKa = 3.49LSGYY580 pKa = 8.25RR581 pKa = 11.84HH582 pKa = 5.82TMQEE586 pKa = 3.69IAADD590 pKa = 4.0ACIMTQDD597 pKa = 3.08QVGYY601 pKa = 8.81FRR603 pKa = 11.84CEE605 pKa = 3.45QCGKK609 pKa = 9.9VLHH612 pKa = 6.16SQYY615 pKa = 11.34DD616 pKa = 3.93LNEE619 pKa = 3.85HH620 pKa = 6.97RR621 pKa = 11.84ISVHH625 pKa = 3.51KK626 pKa = 9.38WKK628 pKa = 10.21RR629 pKa = 11.84VSKK632 pKa = 10.2GRR634 pKa = 11.84PTGSCSIASEE644 pKa = 4.07IYY646 pKa = 10.23GPKK649 pKa = 9.84EE650 pKa = 3.71NPPKK654 pKa = 10.13EE655 pKa = 4.4EE656 pKa = 4.48KK657 pKa = 10.09PVSQPQRR664 pKa = 11.84NRR666 pKa = 11.84RR667 pKa = 11.84EE668 pKa = 3.85VHH670 pKa = 5.61EE671 pKa = 4.61EE672 pKa = 3.84EE673 pKa = 4.87LEE675 pKa = 3.93KK676 pKa = 10.89ALPYY680 pKa = 10.65KK681 pKa = 10.33CTDD684 pKa = 3.23CNRR687 pKa = 11.84RR688 pKa = 11.84FATEE692 pKa = 3.98SARR695 pKa = 11.84LAHH698 pKa = 6.79FGAVHH703 pKa = 7.57AIHH706 pKa = 7.13IAPEE710 pKa = 4.13SAIPTDD716 pKa = 3.34SQDD719 pKa = 3.29PVKK722 pKa = 10.14TDD724 pKa = 3.29RR725 pKa = 11.84AFLGQRR731 pKa = 11.84SRR733 pKa = 11.84LRR735 pKa = 11.84KK736 pKa = 8.76QRR738 pKa = 11.84RR739 pKa = 11.84TKK741 pKa = 10.77SADD744 pKa = 3.21TSVSLAGQTLLQSLQAGQSEE764 pKa = 4.43ISHH767 pKa = 6.77CLQSVSRR774 pKa = 11.84SLNEE778 pKa = 3.75LQKK781 pKa = 10.83VIHH784 pKa = 6.24GRR786 pKa = 11.84NSGTTQKK793 pKa = 11.33
MM1 pKa = 7.66LNALWNQRR9 pKa = 11.84KK10 pKa = 9.09LFVVGITLYY19 pKa = 10.9LVLSGWSGLPLHH31 pKa = 6.04TLVFRR36 pKa = 11.84TVRR39 pKa = 11.84YY40 pKa = 8.85LVWKK44 pKa = 9.9YY45 pKa = 9.02VLRR48 pKa = 11.84DD49 pKa = 4.05TIAGQFLEE57 pKa = 4.43EE58 pKa = 4.58LSSEE62 pKa = 4.13ILGKK66 pKa = 10.27EE67 pKa = 4.01MPPPPPPPPPKK78 pKa = 9.96PIPPLTDD85 pKa = 2.82RR86 pKa = 11.84LYY88 pKa = 10.88EE89 pKa = 4.04DD90 pKa = 3.97WIGLYY95 pKa = 10.55GRR97 pKa = 11.84TVNSTTRR104 pKa = 11.84VLEE107 pKa = 3.8KK108 pKa = 10.28AYY110 pKa = 10.48YY111 pKa = 10.21SSWTSWFHH119 pKa = 5.09MACNTIAIIVAIRR132 pKa = 11.84TLFVAISRR140 pKa = 11.84IIPGSPRR147 pKa = 11.84TAVATWAKK155 pKa = 10.01CKK157 pKa = 10.04IRR159 pKa = 11.84GIHH162 pKa = 5.8YY163 pKa = 7.45EE164 pKa = 3.77AAIQGSLLRR173 pKa = 11.84KK174 pKa = 8.41ATIPEE179 pKa = 3.99WQVEE183 pKa = 4.3VLTPGFLSDD192 pKa = 3.23THH194 pKa = 7.11VGWGVRR200 pKa = 11.84VVDD203 pKa = 3.71TLVVPTHH210 pKa = 5.88VLVEE214 pKa = 4.43AGPNYY219 pKa = 10.21LLKK222 pKa = 10.91GRR224 pKa = 11.84TGSVLCNGLPRR235 pKa = 11.84DD236 pKa = 3.81SRR238 pKa = 11.84IMQDD242 pKa = 2.42ISYY245 pKa = 10.39VPVEE249 pKa = 3.82VSTWSKK255 pKa = 11.05LGTPSVKK262 pKa = 9.9RR263 pKa = 11.84KK264 pKa = 10.08RR265 pKa = 11.84IDD267 pKa = 3.25VEE269 pKa = 4.51TVQSVSCCGLEE280 pKa = 4.33GASTGPLRR288 pKa = 11.84QTRR291 pKa = 11.84VRR293 pKa = 11.84GLMYY297 pKa = 10.83YY298 pKa = 10.1EE299 pKa = 4.59GTTMPGMSGAGYY311 pKa = 10.51YY312 pKa = 9.7IGNTCLGIHH321 pKa = 6.85LGASTNVNCGATIMVPFAEE340 pKa = 4.7IPAPNNLVVQVEE352 pKa = 4.81RR353 pKa = 11.84SAKK356 pKa = 9.56RR357 pKa = 11.84PKK359 pKa = 9.1KK360 pKa = 8.51TRR362 pKa = 11.84PPSGTSADD370 pKa = 3.89PTYY373 pKa = 11.02HH374 pKa = 7.78AMDD377 pKa = 3.57NTKK380 pKa = 8.8TAWGYY385 pKa = 11.02ADD387 pKa = 3.68IRR389 pKa = 11.84EE390 pKa = 4.4ALDD393 pKa = 4.1DD394 pKa = 3.89YY395 pKa = 11.73DD396 pKa = 5.56RR397 pKa = 11.84FIGPSDD403 pKa = 3.48WNNPTDD409 pKa = 5.34DD410 pKa = 4.12FDD412 pKa = 5.13YY413 pKa = 11.27DD414 pKa = 3.17QDD416 pKa = 3.59LGYY419 pKa = 10.6EE420 pKa = 4.23YY421 pKa = 10.62EE422 pKa = 4.3EE423 pKa = 4.27EE424 pKa = 3.92AATRR428 pKa = 11.84GNTLMEE434 pKa = 4.49QIRR437 pKa = 11.84RR438 pKa = 11.84MSVEE442 pKa = 4.26DD443 pKa = 3.28RR444 pKa = 11.84KK445 pKa = 11.09LWMDD449 pKa = 4.6SINAVATEE457 pKa = 4.0EE458 pKa = 4.06AVAARR463 pKa = 11.84STPVANPLPSVRR475 pKa = 11.84PMVQTRR481 pKa = 11.84QGVPRR486 pKa = 11.84QVTFDD491 pKa = 3.86EE492 pKa = 5.24IIVHH496 pKa = 5.62NQTGVGTSTTIHH508 pKa = 6.0TQPQPQRR515 pKa = 11.84LAGPPHH521 pKa = 7.12LGDD524 pKa = 4.74LVRR527 pKa = 11.84DD528 pKa = 4.22LIKK531 pKa = 10.51RR532 pKa = 11.84VTILEE537 pKa = 3.86QKK539 pKa = 9.76MIQRR543 pKa = 11.84DD544 pKa = 3.42EE545 pKa = 4.8RR546 pKa = 11.84MVEE549 pKa = 3.6IEE551 pKa = 4.15NAVGTLQDD559 pKa = 3.45AVAKK563 pKa = 10.01YY564 pKa = 9.93GSPEE568 pKa = 3.84EE569 pKa = 4.16LEE571 pKa = 4.15KK572 pKa = 11.12LRR574 pKa = 11.84NDD576 pKa = 3.49LSGYY580 pKa = 8.25RR581 pKa = 11.84HH582 pKa = 5.82TMQEE586 pKa = 3.69IAADD590 pKa = 4.0ACIMTQDD597 pKa = 3.08QVGYY601 pKa = 8.81FRR603 pKa = 11.84CEE605 pKa = 3.45QCGKK609 pKa = 9.9VLHH612 pKa = 6.16SQYY615 pKa = 11.34DD616 pKa = 3.93LNEE619 pKa = 3.85HH620 pKa = 6.97RR621 pKa = 11.84ISVHH625 pKa = 3.51KK626 pKa = 9.38WKK628 pKa = 10.21RR629 pKa = 11.84VSKK632 pKa = 10.2GRR634 pKa = 11.84PTGSCSIASEE644 pKa = 4.07IYY646 pKa = 10.23GPKK649 pKa = 9.84EE650 pKa = 3.71NPPKK654 pKa = 10.13EE655 pKa = 4.4EE656 pKa = 4.48KK657 pKa = 10.09PVSQPQRR664 pKa = 11.84NRR666 pKa = 11.84RR667 pKa = 11.84EE668 pKa = 3.85VHH670 pKa = 5.61EE671 pKa = 4.61EE672 pKa = 3.84EE673 pKa = 4.87LEE675 pKa = 3.93KK676 pKa = 10.89ALPYY680 pKa = 10.65KK681 pKa = 10.33CTDD684 pKa = 3.23CNRR687 pKa = 11.84RR688 pKa = 11.84FATEE692 pKa = 3.98SARR695 pKa = 11.84LAHH698 pKa = 6.79FGAVHH703 pKa = 7.57AIHH706 pKa = 7.13IAPEE710 pKa = 4.13SAIPTDD716 pKa = 3.34SQDD719 pKa = 3.29PVKK722 pKa = 10.14TDD724 pKa = 3.29RR725 pKa = 11.84AFLGQRR731 pKa = 11.84SRR733 pKa = 11.84LRR735 pKa = 11.84KK736 pKa = 8.76QRR738 pKa = 11.84RR739 pKa = 11.84TKK741 pKa = 10.77SADD744 pKa = 3.21TSVSLAGQTLLQSLQAGQSEE764 pKa = 4.43ISHH767 pKa = 6.77CLQSVSRR774 pKa = 11.84SLNEE778 pKa = 3.75LQKK781 pKa = 10.83VIHH784 pKa = 6.24GRR786 pKa = 11.84NSGTTQKK793 pKa = 11.33
Molecular weight: 88.51 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1190 |
397 |
793 |
595.0 |
66.99 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.723 ± 0.095 | 2.017 ± 0.276 |
4.79 ± 0.275 | 6.723 ± 0.597 |
2.353 ± 0.645 | 6.975 ± 0.181 |
2.605 ± 0.091 | 6.303 ± 0.828 |
4.538 ± 0.136 | 8.151 ± 0.089 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.269 ± 0.137 | 3.193 ± 0.094 |
6.134 ± 0.603 | 4.202 ± 0.51 |
7.143 ± 0.326 | 6.218 ± 0.787 |
7.227 ± 0.372 | 6.723 ± 1.203 |
2.101 ± 0.368 | 3.613 ± 0.367 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
