
Salmonella phage Astrid
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Podoviridae; Astrithrvirus; unclassified Astrithrvirus
Average proteome isoelectric point is 6.02
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 15 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
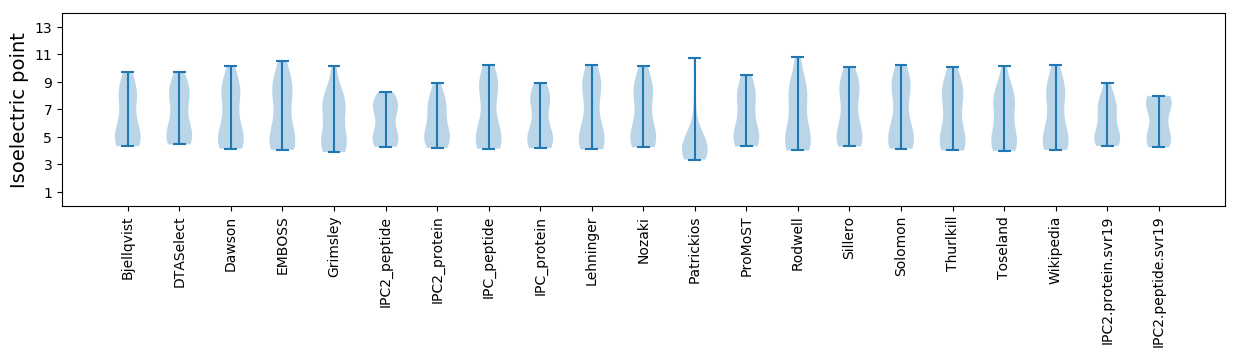
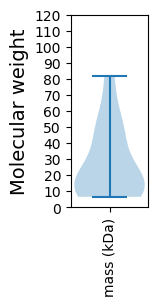
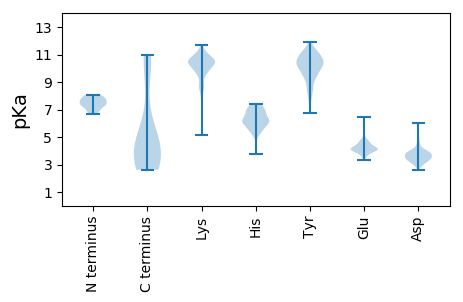
Protein with the lowest isoelectric point:
>tr|A0A3G8F2F9|A0A3G8F2F9_9CAUD Uncharacterized protein OS=Salmonella phage Astrid OX=2483851 PE=4 SV=1
MM1 pKa = 7.36ANVYY5 pKa = 10.32RR6 pKa = 11.84FTQADD11 pKa = 3.05IDD13 pKa = 3.7AAAAQGVTMKK23 pKa = 10.95LSGLDD28 pKa = 3.3VSDD31 pKa = 4.56FDD33 pKa = 5.05GFSDD37 pKa = 4.6SDD39 pKa = 4.01TLVATASGGRR49 pKa = 11.84KK50 pKa = 7.57FTVIPASGFDD60 pKa = 3.5PAYY63 pKa = 8.61TSIYY67 pKa = 9.53FFGWNPSEE75 pKa = 4.07GEE77 pKa = 3.92NMNISFVLSNNDD89 pKa = 2.7TVATLKK95 pKa = 10.52PAVGMNFTKK104 pKa = 10.46FFVTTQAAAVALAWVNHH121 pKa = 6.23PSSGSEE127 pKa = 3.51NTDD130 pKa = 3.29IVFSWDD136 pKa = 3.07GGTLTTGNYY145 pKa = 9.52NVVVTKK151 pKa = 10.86DD152 pKa = 3.04GAEE155 pKa = 4.09VVNISTPDD163 pKa = 3.05KK164 pKa = 11.13SYY166 pKa = 9.62TLNKK170 pKa = 8.68TAGDD174 pKa = 4.1YY175 pKa = 11.08VVTVYY180 pKa = 11.22DD181 pKa = 3.91KK182 pKa = 11.42GGKK185 pKa = 9.17SDD187 pKa = 3.65STASNISQAITLSAVLPKK205 pKa = 10.6ALYY208 pKa = 8.3TVKK211 pKa = 10.62AADD214 pKa = 3.16ITAISDD220 pKa = 3.5NEE222 pKa = 3.46IDD224 pKa = 3.77MKK226 pKa = 11.67VNGADD231 pKa = 2.97ITAGSVLRR239 pKa = 11.84LGDD242 pKa = 3.65VIVAKK247 pKa = 10.85VSGIRR252 pKa = 11.84KK253 pKa = 9.29FYY255 pKa = 10.85TDD257 pKa = 3.33TTLHH261 pKa = 5.37GTSINFAVFFDD272 pKa = 5.81GEE274 pKa = 4.65SQWLQFALSDD284 pKa = 3.71NDD286 pKa = 3.37QTATFTMVDD295 pKa = 3.97DD296 pKa = 4.42TSGSAGTYY304 pKa = 6.73QAWNIRR310 pKa = 11.84TKK312 pKa = 10.57QEE314 pKa = 3.66TPAVVGTNNVYY325 pKa = 10.66KK326 pKa = 9.93IDD328 pKa = 3.8TSILSSVNKK337 pKa = 9.76EE338 pKa = 3.73RR339 pKa = 11.84FVTITGSDD347 pKa = 3.39TPFDD351 pKa = 3.8YY352 pKa = 10.84GQYY355 pKa = 9.88ILSVLQFPFDD365 pKa = 3.58IPADD369 pKa = 3.67QILTPEE375 pKa = 4.73NIQLANRR382 pKa = 11.84QLSVAANKK390 pKa = 9.81VATDD394 pKa = 4.32KK395 pKa = 11.22IKK397 pKa = 10.42IDD399 pKa = 3.4LGEE402 pKa = 4.01IVVPDD407 pKa = 3.77TYY409 pKa = 11.91GNMLSFANTNAVIHH423 pKa = 6.43LPLAPSIVLDD433 pKa = 3.7LEE435 pKa = 4.46YY436 pKa = 11.1VIGQTLGVYY445 pKa = 10.19YY446 pKa = 10.95LLDD449 pKa = 4.86CYY451 pKa = 10.79TGTATINITSTKK463 pKa = 8.71LAAVISSTQVNIGVRR478 pKa = 11.84VPYY481 pKa = 9.89MADD484 pKa = 2.97SYY486 pKa = 10.14TAPEE490 pKa = 4.06NTGVVAGGNNGVKK503 pKa = 9.69IPYY506 pKa = 9.48IEE508 pKa = 5.51LISHH512 pKa = 7.26DD513 pKa = 5.09AILPHH518 pKa = 6.82GFFTVPVVDD527 pKa = 3.68EE528 pKa = 4.36TLISGQTGYY537 pKa = 10.64IKK539 pKa = 10.53VDD541 pKa = 3.2NVEE544 pKa = 4.44LVTGALGNEE553 pKa = 4.02KK554 pKa = 10.56AQIISLLNSGVIIKK568 pKa = 10.32
MM1 pKa = 7.36ANVYY5 pKa = 10.32RR6 pKa = 11.84FTQADD11 pKa = 3.05IDD13 pKa = 3.7AAAAQGVTMKK23 pKa = 10.95LSGLDD28 pKa = 3.3VSDD31 pKa = 4.56FDD33 pKa = 5.05GFSDD37 pKa = 4.6SDD39 pKa = 4.01TLVATASGGRR49 pKa = 11.84KK50 pKa = 7.57FTVIPASGFDD60 pKa = 3.5PAYY63 pKa = 8.61TSIYY67 pKa = 9.53FFGWNPSEE75 pKa = 4.07GEE77 pKa = 3.92NMNISFVLSNNDD89 pKa = 2.7TVATLKK95 pKa = 10.52PAVGMNFTKK104 pKa = 10.46FFVTTQAAAVALAWVNHH121 pKa = 6.23PSSGSEE127 pKa = 3.51NTDD130 pKa = 3.29IVFSWDD136 pKa = 3.07GGTLTTGNYY145 pKa = 9.52NVVVTKK151 pKa = 10.86DD152 pKa = 3.04GAEE155 pKa = 4.09VVNISTPDD163 pKa = 3.05KK164 pKa = 11.13SYY166 pKa = 9.62TLNKK170 pKa = 8.68TAGDD174 pKa = 4.1YY175 pKa = 11.08VVTVYY180 pKa = 11.22DD181 pKa = 3.91KK182 pKa = 11.42GGKK185 pKa = 9.17SDD187 pKa = 3.65STASNISQAITLSAVLPKK205 pKa = 10.6ALYY208 pKa = 8.3TVKK211 pKa = 10.62AADD214 pKa = 3.16ITAISDD220 pKa = 3.5NEE222 pKa = 3.46IDD224 pKa = 3.77MKK226 pKa = 11.67VNGADD231 pKa = 2.97ITAGSVLRR239 pKa = 11.84LGDD242 pKa = 3.65VIVAKK247 pKa = 10.85VSGIRR252 pKa = 11.84KK253 pKa = 9.29FYY255 pKa = 10.85TDD257 pKa = 3.33TTLHH261 pKa = 5.37GTSINFAVFFDD272 pKa = 5.81GEE274 pKa = 4.65SQWLQFALSDD284 pKa = 3.71NDD286 pKa = 3.37QTATFTMVDD295 pKa = 3.97DD296 pKa = 4.42TSGSAGTYY304 pKa = 6.73QAWNIRR310 pKa = 11.84TKK312 pKa = 10.57QEE314 pKa = 3.66TPAVVGTNNVYY325 pKa = 10.66KK326 pKa = 9.93IDD328 pKa = 3.8TSILSSVNKK337 pKa = 9.76EE338 pKa = 3.73RR339 pKa = 11.84FVTITGSDD347 pKa = 3.39TPFDD351 pKa = 3.8YY352 pKa = 10.84GQYY355 pKa = 9.88ILSVLQFPFDD365 pKa = 3.58IPADD369 pKa = 3.67QILTPEE375 pKa = 4.73NIQLANRR382 pKa = 11.84QLSVAANKK390 pKa = 9.81VATDD394 pKa = 4.32KK395 pKa = 11.22IKK397 pKa = 10.42IDD399 pKa = 3.4LGEE402 pKa = 4.01IVVPDD407 pKa = 3.77TYY409 pKa = 11.91GNMLSFANTNAVIHH423 pKa = 6.43LPLAPSIVLDD433 pKa = 3.7LEE435 pKa = 4.46YY436 pKa = 11.1VIGQTLGVYY445 pKa = 10.19YY446 pKa = 10.95LLDD449 pKa = 4.86CYY451 pKa = 10.79TGTATINITSTKK463 pKa = 8.71LAAVISSTQVNIGVRR478 pKa = 11.84VPYY481 pKa = 9.89MADD484 pKa = 2.97SYY486 pKa = 10.14TAPEE490 pKa = 4.06NTGVVAGGNNGVKK503 pKa = 9.69IPYY506 pKa = 9.48IEE508 pKa = 5.51LISHH512 pKa = 7.26DD513 pKa = 5.09AILPHH518 pKa = 6.82GFFTVPVVDD527 pKa = 3.68EE528 pKa = 4.36TLISGQTGYY537 pKa = 10.64IKK539 pKa = 10.53VDD541 pKa = 3.2NVEE544 pKa = 4.44LVTGALGNEE553 pKa = 4.02KK554 pKa = 10.56AQIISLLNSGVIIKK568 pKa = 10.32
Molecular weight: 60.57 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3G8F4Y3|A0A3G8F4Y3_9CAUD DNA-directed DNA polymerase OS=Salmonella phage Astrid OX=2483851 PE=4 SV=1
MM1 pKa = 7.88GINITRR7 pKa = 11.84QRR9 pKa = 11.84LLDD12 pKa = 3.53GMRR15 pKa = 11.84SIGEE19 pKa = 4.23PFTQSEE25 pKa = 4.33IEE27 pKa = 4.47SICRR31 pKa = 11.84EE32 pKa = 4.01LFHH35 pKa = 7.08NRR37 pKa = 11.84GKK39 pKa = 10.51ISDD42 pKa = 4.49LKK44 pKa = 11.19NKK46 pKa = 9.57LASKK50 pKa = 10.39SDD52 pKa = 3.5NKK54 pKa = 10.32KK55 pKa = 10.83APP57 pKa = 3.43
MM1 pKa = 7.88GINITRR7 pKa = 11.84QRR9 pKa = 11.84LLDD12 pKa = 3.53GMRR15 pKa = 11.84SIGEE19 pKa = 4.23PFTQSEE25 pKa = 4.33IEE27 pKa = 4.47SICRR31 pKa = 11.84EE32 pKa = 4.01LFHH35 pKa = 7.08NRR37 pKa = 11.84GKK39 pKa = 10.51ISDD42 pKa = 4.49LKK44 pKa = 11.19NKK46 pKa = 9.57LASKK50 pKa = 10.39SDD52 pKa = 3.5NKK54 pKa = 10.32KK55 pKa = 10.83APP57 pKa = 3.43
Molecular weight: 6.47 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3632 |
57 |
708 |
242.1 |
27.19 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.828 ± 0.805 | 0.798 ± 0.222 |
6.635 ± 0.403 | 6.47 ± 0.8 |
4.268 ± 0.249 | 6.333 ± 0.396 |
1.377 ± 0.319 | 6.718 ± 0.361 |
7.461 ± 0.725 | 7.764 ± 0.429 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.029 ± 0.347 | 6.856 ± 0.639 |
2.671 ± 0.248 | 3.139 ± 0.595 |
3.111 ± 0.514 | 6.525 ± 0.417 |
7.048 ± 0.782 | 7.241 ± 0.818 |
0.964 ± 0.132 | 4.763 ± 0.784 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
