
Agromyces sp. SJ-23
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Agromyces
Average proteome isoelectric point is 5.92
Get precalculated fractions of proteins
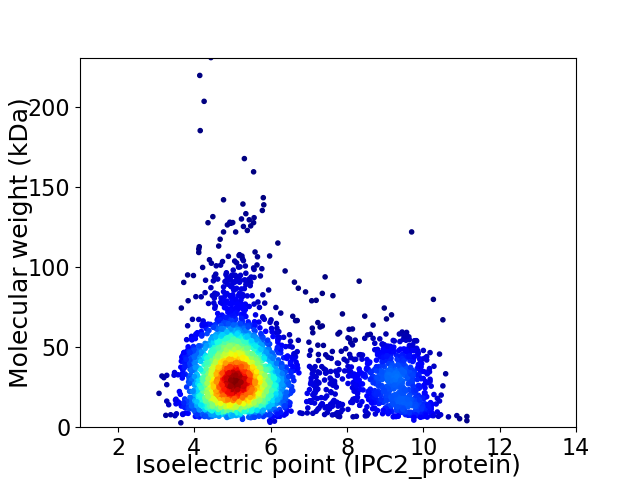
Virtual 2D-PAGE plot for 3729 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
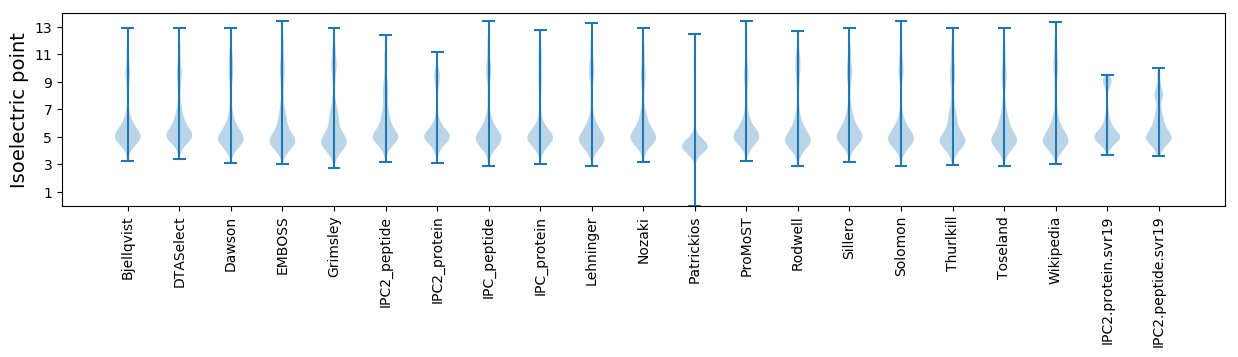
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3M7ZVZ5|A0A3M7ZVZ5_9MICO ATP-binding protein (Fragment) OS=Agromyces sp. SJ-23 OX=2583849 GN=EDM22_19280 PE=4 SV=1
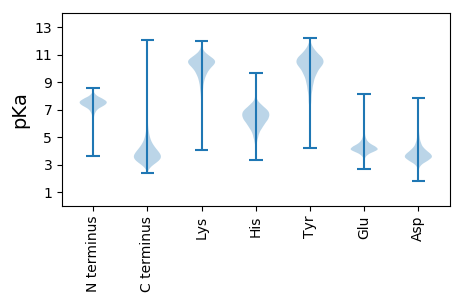
MM1 pKa = 8.04RR2 pKa = 11.84FPIHH6 pKa = 6.33VPAAVDD12 pKa = 3.32DD13 pKa = 3.83RR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84PVRR19 pKa = 11.84LRR21 pKa = 11.84LAALGVATILAAGGAFATPFAAHH44 pKa = 6.82AEE46 pKa = 4.23EE47 pKa = 4.41AQPAGGEE54 pKa = 4.05SAMLVEE60 pKa = 5.12AAASEE65 pKa = 4.28PVAAEE70 pKa = 4.19DD71 pKa = 4.38PAPADD76 pKa = 3.58EE77 pKa = 5.19GEE79 pKa = 4.47TGASATEE86 pKa = 4.14PAASATDD93 pKa = 3.88DD94 pKa = 4.13DD95 pKa = 4.95AATAHH100 pKa = 7.01ADD102 pKa = 3.47AAADD106 pKa = 3.75PAGPAAGAAGDD117 pKa = 4.41PEE119 pKa = 5.24ASHH122 pKa = 6.94LPDD125 pKa = 5.0AEE127 pKa = 4.39AEE129 pKa = 4.08PVAARR134 pKa = 11.84KK135 pKa = 9.06SAASAGQPAGFAAQVEE151 pKa = 4.45PSQLGPNQAPIAVDD165 pKa = 3.48DD166 pKa = 4.94AYY168 pKa = 11.88QMLADD173 pKa = 3.86TTLTVDD179 pKa = 4.1DD180 pKa = 5.21PGVTGNDD187 pKa = 3.72TEE189 pKa = 5.22PDD191 pKa = 3.7GDD193 pKa = 3.61WYY195 pKa = 11.18QVDD198 pKa = 4.63DD199 pKa = 3.83HH200 pKa = 6.8TKK202 pKa = 9.07PQVGSLWVNAHH213 pKa = 6.08GPFGYY218 pKa = 8.84TPPAGFTGTVTFTYY232 pKa = 10.6VLKK235 pKa = 10.74DD236 pKa = 3.44DD237 pKa = 5.17FGALSTWATVTIEE250 pKa = 3.96VLPAGSDD257 pKa = 3.08ATLPPTANDD266 pKa = 3.32DD267 pKa = 4.02TYY269 pKa = 11.75VYY271 pKa = 11.15ALSTPLYY278 pKa = 8.49IAAPGVLANDD288 pKa = 3.93DD289 pKa = 3.76AEE291 pKa = 4.38GRR293 pKa = 11.84EE294 pKa = 4.37GTLVLDD300 pKa = 4.06YY301 pKa = 10.89ASPQIGTVDD310 pKa = 3.92LQADD314 pKa = 4.26GSFLYY319 pKa = 10.01TPHH322 pKa = 7.55DD323 pKa = 4.84SIGGTWWFRR332 pKa = 11.84YY333 pKa = 8.86LLCTDD338 pKa = 4.54GGCASAEE345 pKa = 4.18VTLEE349 pKa = 3.57QAAAGEE355 pKa = 4.43QPSGPSQPPAEE366 pKa = 4.5PPAGEE371 pKa = 4.52DD372 pKa = 3.61LAPVAQPDD380 pKa = 3.88TLVAVAGDD388 pKa = 4.09LAILDD393 pKa = 4.01APGVLGNDD401 pKa = 4.01SDD403 pKa = 5.59PEE405 pKa = 4.3GQPLTLVDD413 pKa = 3.84VTTPAHH419 pKa = 5.44GVLYY423 pKa = 10.11YY424 pKa = 10.01WNADD428 pKa = 3.1GTVQYY433 pKa = 10.94VPNDD437 pKa = 3.38GFAGTDD443 pKa = 3.21QVEE446 pKa = 4.49YY447 pKa = 9.89TVSDD451 pKa = 3.98GAKK454 pKa = 9.85SATSTLTFTVTEE466 pKa = 4.13PANYY470 pKa = 9.98APQAWEE476 pKa = 4.1DD477 pKa = 3.74SAVAVAGTTLILDD490 pKa = 3.93APGVLGNDD498 pKa = 3.59SDD500 pKa = 4.62QDD502 pKa = 3.67GDD504 pKa = 4.01PLAVTWFGDD513 pKa = 3.64PHH515 pKa = 7.82HH516 pKa = 6.38GTIDD520 pKa = 3.13IAADD524 pKa = 3.64GSVVYY529 pKa = 9.65TPEE532 pKa = 5.05AGFVGGDD539 pKa = 3.34SVLYY543 pKa = 9.82QASDD547 pKa = 3.53GQATSEE553 pKa = 4.15AFLVIDD559 pKa = 4.24VVSAGAPAHH568 pKa = 5.38PTVVGDD574 pKa = 4.08HH575 pKa = 6.6YY576 pKa = 11.44DD577 pKa = 3.34AVSGVLLEE585 pKa = 4.31VSAPGVLGNDD595 pKa = 4.53LDD597 pKa = 4.19PSGPIAVTGHH607 pKa = 6.21EE608 pKa = 4.24AAQHH612 pKa = 4.41GTIEE616 pKa = 4.06IAADD620 pKa = 3.51GSLRR624 pKa = 11.84YY625 pKa = 8.14TSEE628 pKa = 3.82PGYY631 pKa = 9.62TGVDD635 pKa = 3.85TIRR638 pKa = 11.84YY639 pKa = 5.92TISDD643 pKa = 4.09GEE645 pKa = 4.12EE646 pKa = 4.05SADD649 pKa = 3.42GLISITVLGQAEE661 pKa = 4.8GSGQPGEE668 pKa = 4.5GEE670 pKa = 4.11QPGGSGGSGGPQQPGRR686 pKa = 11.84PQQPTGPDD694 pKa = 3.25EE695 pKa = 4.51PGRR698 pKa = 11.84PAEE701 pKa = 4.17HH702 pKa = 6.86APLPEE707 pKa = 4.47ARR709 pKa = 11.84PATTDD714 pKa = 3.11AAALADD720 pKa = 3.83TGSDD724 pKa = 3.32PRR726 pKa = 11.84LAAALAALLAALGII740 pKa = 4.44
MM1 pKa = 8.04RR2 pKa = 11.84FPIHH6 pKa = 6.33VPAAVDD12 pKa = 3.32DD13 pKa = 3.83RR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84PVRR19 pKa = 11.84LRR21 pKa = 11.84LAALGVATILAAGGAFATPFAAHH44 pKa = 6.82AEE46 pKa = 4.23EE47 pKa = 4.41AQPAGGEE54 pKa = 4.05SAMLVEE60 pKa = 5.12AAASEE65 pKa = 4.28PVAAEE70 pKa = 4.19DD71 pKa = 4.38PAPADD76 pKa = 3.58EE77 pKa = 5.19GEE79 pKa = 4.47TGASATEE86 pKa = 4.14PAASATDD93 pKa = 3.88DD94 pKa = 4.13DD95 pKa = 4.95AATAHH100 pKa = 7.01ADD102 pKa = 3.47AAADD106 pKa = 3.75PAGPAAGAAGDD117 pKa = 4.41PEE119 pKa = 5.24ASHH122 pKa = 6.94LPDD125 pKa = 5.0AEE127 pKa = 4.39AEE129 pKa = 4.08PVAARR134 pKa = 11.84KK135 pKa = 9.06SAASAGQPAGFAAQVEE151 pKa = 4.45PSQLGPNQAPIAVDD165 pKa = 3.48DD166 pKa = 4.94AYY168 pKa = 11.88QMLADD173 pKa = 3.86TTLTVDD179 pKa = 4.1DD180 pKa = 5.21PGVTGNDD187 pKa = 3.72TEE189 pKa = 5.22PDD191 pKa = 3.7GDD193 pKa = 3.61WYY195 pKa = 11.18QVDD198 pKa = 4.63DD199 pKa = 3.83HH200 pKa = 6.8TKK202 pKa = 9.07PQVGSLWVNAHH213 pKa = 6.08GPFGYY218 pKa = 8.84TPPAGFTGTVTFTYY232 pKa = 10.6VLKK235 pKa = 10.74DD236 pKa = 3.44DD237 pKa = 5.17FGALSTWATVTIEE250 pKa = 3.96VLPAGSDD257 pKa = 3.08ATLPPTANDD266 pKa = 3.32DD267 pKa = 4.02TYY269 pKa = 11.75VYY271 pKa = 11.15ALSTPLYY278 pKa = 8.49IAAPGVLANDD288 pKa = 3.93DD289 pKa = 3.76AEE291 pKa = 4.38GRR293 pKa = 11.84EE294 pKa = 4.37GTLVLDD300 pKa = 4.06YY301 pKa = 10.89ASPQIGTVDD310 pKa = 3.92LQADD314 pKa = 4.26GSFLYY319 pKa = 10.01TPHH322 pKa = 7.55DD323 pKa = 4.84SIGGTWWFRR332 pKa = 11.84YY333 pKa = 8.86LLCTDD338 pKa = 4.54GGCASAEE345 pKa = 4.18VTLEE349 pKa = 3.57QAAAGEE355 pKa = 4.43QPSGPSQPPAEE366 pKa = 4.5PPAGEE371 pKa = 4.52DD372 pKa = 3.61LAPVAQPDD380 pKa = 3.88TLVAVAGDD388 pKa = 4.09LAILDD393 pKa = 4.01APGVLGNDD401 pKa = 4.01SDD403 pKa = 5.59PEE405 pKa = 4.3GQPLTLVDD413 pKa = 3.84VTTPAHH419 pKa = 5.44GVLYY423 pKa = 10.11YY424 pKa = 10.01WNADD428 pKa = 3.1GTVQYY433 pKa = 10.94VPNDD437 pKa = 3.38GFAGTDD443 pKa = 3.21QVEE446 pKa = 4.49YY447 pKa = 9.89TVSDD451 pKa = 3.98GAKK454 pKa = 9.85SATSTLTFTVTEE466 pKa = 4.13PANYY470 pKa = 9.98APQAWEE476 pKa = 4.1DD477 pKa = 3.74SAVAVAGTTLILDD490 pKa = 3.93APGVLGNDD498 pKa = 3.59SDD500 pKa = 4.62QDD502 pKa = 3.67GDD504 pKa = 4.01PLAVTWFGDD513 pKa = 3.64PHH515 pKa = 7.82HH516 pKa = 6.38GTIDD520 pKa = 3.13IAADD524 pKa = 3.64GSVVYY529 pKa = 9.65TPEE532 pKa = 5.05AGFVGGDD539 pKa = 3.34SVLYY543 pKa = 9.82QASDD547 pKa = 3.53GQATSEE553 pKa = 4.15AFLVIDD559 pKa = 4.24VVSAGAPAHH568 pKa = 5.38PTVVGDD574 pKa = 4.08HH575 pKa = 6.6YY576 pKa = 11.44DD577 pKa = 3.34AVSGVLLEE585 pKa = 4.31VSAPGVLGNDD595 pKa = 4.53LDD597 pKa = 4.19PSGPIAVTGHH607 pKa = 6.21EE608 pKa = 4.24AAQHH612 pKa = 4.41GTIEE616 pKa = 4.06IAADD620 pKa = 3.51GSLRR624 pKa = 11.84YY625 pKa = 8.14TSEE628 pKa = 3.82PGYY631 pKa = 9.62TGVDD635 pKa = 3.85TIRR638 pKa = 11.84YY639 pKa = 5.92TISDD643 pKa = 4.09GEE645 pKa = 4.12EE646 pKa = 4.05SADD649 pKa = 3.42GLISITVLGQAEE661 pKa = 4.8GSGQPGEE668 pKa = 4.5GEE670 pKa = 4.11QPGGSGGSGGPQQPGRR686 pKa = 11.84PQQPTGPDD694 pKa = 3.25EE695 pKa = 4.51PGRR698 pKa = 11.84PAEE701 pKa = 4.17HH702 pKa = 6.86APLPEE707 pKa = 4.47ARR709 pKa = 11.84PATTDD714 pKa = 3.11AAALADD720 pKa = 3.83TGSDD724 pKa = 3.32PRR726 pKa = 11.84LAAALAALLAALGII740 pKa = 4.44
Molecular weight: 74.49 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3M8A883|A0A3M8A883_9MICO PPOX class F420-dependent oxidoreductase OS=Agromyces sp. SJ-23 OX=2583849 GN=EDM22_12195 PE=4 SV=1
MM1 pKa = 7.96PLRR4 pKa = 11.84AARR7 pKa = 11.84VGRR10 pKa = 11.84VGVVGAPVAKK20 pKa = 10.13AAVVGAAVSPGRR32 pKa = 11.84SPVAKK37 pKa = 9.86AAVVGAAVTPGRR49 pKa = 11.84SPVAKK54 pKa = 9.86AAVVGAAVTPGRR66 pKa = 11.84RR67 pKa = 11.84RR68 pKa = 11.84RR69 pKa = 11.84II70 pKa = 3.53
MM1 pKa = 7.96PLRR4 pKa = 11.84AARR7 pKa = 11.84VGRR10 pKa = 11.84VGVVGAPVAKK20 pKa = 10.13AAVVGAAVSPGRR32 pKa = 11.84SPVAKK37 pKa = 9.86AAVVGAAVTPGRR49 pKa = 11.84SPVAKK54 pKa = 9.86AAVVGAAVTPGRR66 pKa = 11.84RR67 pKa = 11.84RR68 pKa = 11.84RR69 pKa = 11.84II70 pKa = 3.53
Molecular weight: 6.69 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1218308 |
29 |
2207 |
326.7 |
34.84 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.335 ± 0.064 | 0.467 ± 0.009 |
6.257 ± 0.03 | 5.824 ± 0.03 |
3.153 ± 0.027 | 9.216 ± 0.033 |
2.005 ± 0.022 | 4.242 ± 0.028 |
1.619 ± 0.026 | 10.123 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.671 ± 0.015 | 1.767 ± 0.02 |
5.599 ± 0.028 | 2.49 ± 0.019 |
7.652 ± 0.052 | 5.295 ± 0.029 |
5.656 ± 0.034 | 9.152 ± 0.034 |
1.538 ± 0.018 | 1.939 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
