
Dragonfly larvae associated circular virus-7
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 7.44
Get precalculated fractions of proteins
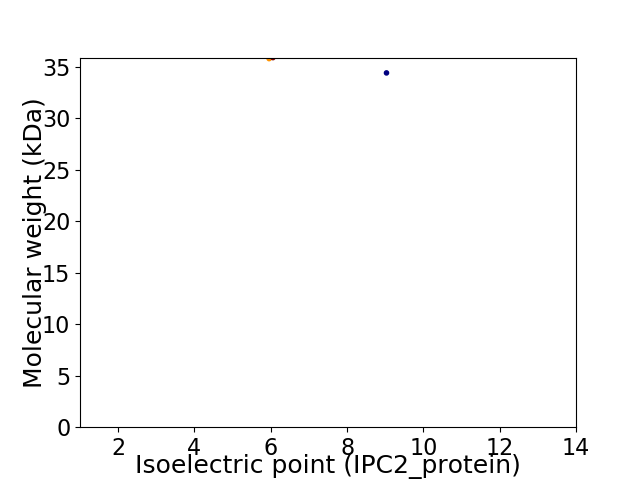
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|W5U7R2|W5U7R2_9VIRU ATP-dependent helicase Rep OS=Dragonfly larvae associated circular virus-7 OX=1454028 PE=3 SV=1
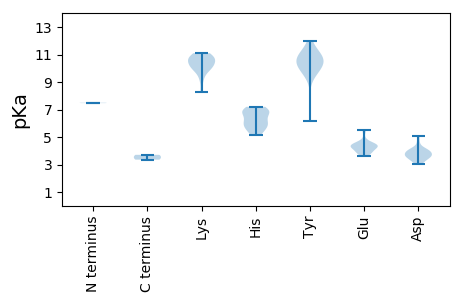
MM1 pKa = 7.5ALILAKK7 pKa = 9.96RR8 pKa = 11.84VRR10 pKa = 11.84RR11 pKa = 11.84VCFTLFEE18 pKa = 4.32EE19 pKa = 4.89VPPEE23 pKa = 4.59FEE25 pKa = 4.44EE26 pKa = 4.54EE27 pKa = 3.73WMQYY31 pKa = 11.06LLVGRR36 pKa = 11.84EE37 pKa = 3.98VCPATGRR44 pKa = 11.84EE45 pKa = 3.86HH46 pKa = 5.13WQGYY50 pKa = 9.93LEE52 pKa = 5.43LKK54 pKa = 10.6SQMLWTTVKK63 pKa = 10.45GRR65 pKa = 11.84LGDD68 pKa = 3.67QVHH71 pKa = 6.83LEE73 pKa = 4.19EE74 pKa = 5.49CNGSAQDD81 pKa = 3.93NIVYY85 pKa = 10.14CSKK88 pKa = 10.78DD89 pKa = 3.64GNWVEE94 pKa = 3.81WGEE97 pKa = 4.44CKK99 pKa = 10.7VQGARR104 pKa = 11.84SDD106 pKa = 3.84LSSLRR111 pKa = 11.84TDD113 pKa = 3.59VLNHH117 pKa = 5.91GVTVAEE123 pKa = 4.04ILEE126 pKa = 4.58ANPMTYY132 pKa = 9.44HH133 pKa = 5.88QYY135 pKa = 11.18GRR137 pKa = 11.84TLLALEE143 pKa = 4.36DD144 pKa = 3.72LRR146 pKa = 11.84YY147 pKa = 10.48SRR149 pKa = 11.84VFRR152 pKa = 11.84DD153 pKa = 4.04CSAAPTVTWFWGPTGCGKK171 pKa = 9.57SHH173 pKa = 6.88EE174 pKa = 4.42SRR176 pKa = 11.84ARR178 pKa = 11.84ALALAGGDD186 pKa = 3.07RR187 pKa = 11.84SKK189 pKa = 10.82IYY191 pKa = 11.0VLANHH196 pKa = 7.2DD197 pKa = 4.0KK198 pKa = 11.04GWWDD202 pKa = 4.0LYY204 pKa = 10.96NLQEE208 pKa = 4.53IVILDD213 pKa = 4.13EE214 pKa = 4.41LRR216 pKa = 11.84ADD218 pKa = 3.66QVPWGLLLQLTTNVEE233 pKa = 4.24CFVPRR238 pKa = 11.84RR239 pKa = 11.84GRR241 pKa = 11.84QPCPFVAKK249 pKa = 10.05HH250 pKa = 5.41VFITSPLHH258 pKa = 6.9PRR260 pKa = 11.84DD261 pKa = 3.61QYY263 pKa = 12.01ADD265 pKa = 3.28RR266 pKa = 11.84CGIDD270 pKa = 5.09NIDD273 pKa = 3.32QLLRR277 pKa = 11.84RR278 pKa = 11.84ITFIEE283 pKa = 4.18EE284 pKa = 3.79RR285 pKa = 11.84NVPYY289 pKa = 10.47AAGVFNHH296 pKa = 6.11SKK298 pKa = 10.72RR299 pKa = 11.84LLFQGSMRR307 pKa = 11.84TEE309 pKa = 3.9RR310 pKa = 11.84EE311 pKa = 3.71
MM1 pKa = 7.5ALILAKK7 pKa = 9.96RR8 pKa = 11.84VRR10 pKa = 11.84RR11 pKa = 11.84VCFTLFEE18 pKa = 4.32EE19 pKa = 4.89VPPEE23 pKa = 4.59FEE25 pKa = 4.44EE26 pKa = 4.54EE27 pKa = 3.73WMQYY31 pKa = 11.06LLVGRR36 pKa = 11.84EE37 pKa = 3.98VCPATGRR44 pKa = 11.84EE45 pKa = 3.86HH46 pKa = 5.13WQGYY50 pKa = 9.93LEE52 pKa = 5.43LKK54 pKa = 10.6SQMLWTTVKK63 pKa = 10.45GRR65 pKa = 11.84LGDD68 pKa = 3.67QVHH71 pKa = 6.83LEE73 pKa = 4.19EE74 pKa = 5.49CNGSAQDD81 pKa = 3.93NIVYY85 pKa = 10.14CSKK88 pKa = 10.78DD89 pKa = 3.64GNWVEE94 pKa = 3.81WGEE97 pKa = 4.44CKK99 pKa = 10.7VQGARR104 pKa = 11.84SDD106 pKa = 3.84LSSLRR111 pKa = 11.84TDD113 pKa = 3.59VLNHH117 pKa = 5.91GVTVAEE123 pKa = 4.04ILEE126 pKa = 4.58ANPMTYY132 pKa = 9.44HH133 pKa = 5.88QYY135 pKa = 11.18GRR137 pKa = 11.84TLLALEE143 pKa = 4.36DD144 pKa = 3.72LRR146 pKa = 11.84YY147 pKa = 10.48SRR149 pKa = 11.84VFRR152 pKa = 11.84DD153 pKa = 4.04CSAAPTVTWFWGPTGCGKK171 pKa = 9.57SHH173 pKa = 6.88EE174 pKa = 4.42SRR176 pKa = 11.84ARR178 pKa = 11.84ALALAGGDD186 pKa = 3.07RR187 pKa = 11.84SKK189 pKa = 10.82IYY191 pKa = 11.0VLANHH196 pKa = 7.2DD197 pKa = 4.0KK198 pKa = 11.04GWWDD202 pKa = 4.0LYY204 pKa = 10.96NLQEE208 pKa = 4.53IVILDD213 pKa = 4.13EE214 pKa = 4.41LRR216 pKa = 11.84ADD218 pKa = 3.66QVPWGLLLQLTTNVEE233 pKa = 4.24CFVPRR238 pKa = 11.84RR239 pKa = 11.84GRR241 pKa = 11.84QPCPFVAKK249 pKa = 10.05HH250 pKa = 5.41VFITSPLHH258 pKa = 6.9PRR260 pKa = 11.84DD261 pKa = 3.61QYY263 pKa = 12.01ADD265 pKa = 3.28RR266 pKa = 11.84CGIDD270 pKa = 5.09NIDD273 pKa = 3.32QLLRR277 pKa = 11.84RR278 pKa = 11.84ITFIEE283 pKa = 4.18EE284 pKa = 3.79RR285 pKa = 11.84NVPYY289 pKa = 10.47AAGVFNHH296 pKa = 6.11SKK298 pKa = 10.72RR299 pKa = 11.84LLFQGSMRR307 pKa = 11.84TEE309 pKa = 3.9RR310 pKa = 11.84EE311 pKa = 3.71
Molecular weight: 35.79 kDa
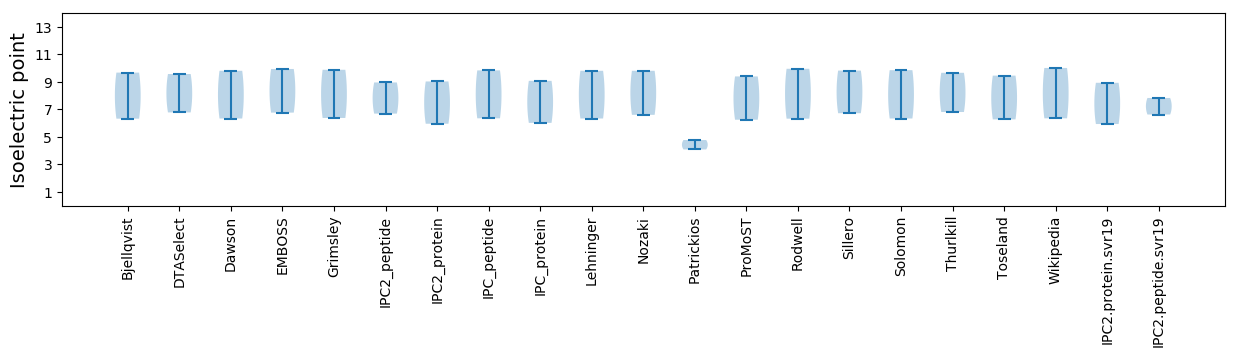
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W5U7R2|W5U7R2_9VIRU ATP-dependent helicase Rep OS=Dragonfly larvae associated circular virus-7 OX=1454028 PE=3 SV=1
MM1 pKa = 7.47SCRR4 pKa = 11.84ACFYY8 pKa = 10.42TFVGRR13 pKa = 11.84NSGKK17 pKa = 9.98VVNSLTLRR25 pKa = 11.84RR26 pKa = 11.84CLRR29 pKa = 11.84VCVCVCVMAWGKK41 pKa = 9.16YY42 pKa = 6.15AHH44 pKa = 6.74RR45 pKa = 11.84RR46 pKa = 11.84HH47 pKa = 5.72YY48 pKa = 9.85GHH50 pKa = 6.98RR51 pKa = 11.84RR52 pKa = 11.84RR53 pKa = 11.84YY54 pKa = 9.78GRR56 pKa = 11.84VNIQRR61 pKa = 11.84NMLGMPSQRR70 pKa = 11.84LVSLRR75 pKa = 11.84QHH77 pKa = 6.28NSWTQRR83 pKa = 11.84MADD86 pKa = 3.72AGSGPAAGTVTMPLLANCPLSITSPLVSGWDD117 pKa = 3.44SDD119 pKa = 4.08PLFWEE124 pKa = 4.43MWSSVYY130 pKa = 10.66SHH132 pKa = 5.62YY133 pKa = 10.18TVVGVKK139 pKa = 8.25FTLKK143 pKa = 10.05TMWAAPTVTPTDD155 pKa = 3.1TEE157 pKa = 4.32GAVRR161 pKa = 11.84VPMRR165 pKa = 11.84VSLMVNTRR173 pKa = 11.84EE174 pKa = 4.3SIPDD178 pKa = 3.46YY179 pKa = 10.84VVPEE183 pKa = 3.9QLQEE187 pKa = 3.62QGLIHH192 pKa = 6.87WKK194 pKa = 10.7DD195 pKa = 3.56LMTSDD200 pKa = 4.62LSRR203 pKa = 11.84VHH205 pKa = 6.22VVEE208 pKa = 4.5KK209 pKa = 10.21FSAKK213 pKa = 10.16KK214 pKa = 10.22FFGLSSVSDD223 pKa = 4.13DD224 pKa = 3.91PSIGAPCTTYY234 pKa = 10.22PAALAWFLILVQKK247 pKa = 11.1VEE249 pKa = 4.15GTPFGGGGEE258 pKa = 4.21AGLPSSCSTVPSSFPSFSATRR279 pKa = 11.84SPRR282 pKa = 11.84PSRR285 pKa = 11.84RR286 pKa = 11.84PRR288 pKa = 11.84QANKK292 pKa = 10.58AIILSLFSCCLEE304 pKa = 4.23KK305 pKa = 10.96GAVYY309 pKa = 10.67YY310 pKa = 10.78GG311 pKa = 3.35
MM1 pKa = 7.47SCRR4 pKa = 11.84ACFYY8 pKa = 10.42TFVGRR13 pKa = 11.84NSGKK17 pKa = 9.98VVNSLTLRR25 pKa = 11.84RR26 pKa = 11.84CLRR29 pKa = 11.84VCVCVCVMAWGKK41 pKa = 9.16YY42 pKa = 6.15AHH44 pKa = 6.74RR45 pKa = 11.84RR46 pKa = 11.84HH47 pKa = 5.72YY48 pKa = 9.85GHH50 pKa = 6.98RR51 pKa = 11.84RR52 pKa = 11.84RR53 pKa = 11.84YY54 pKa = 9.78GRR56 pKa = 11.84VNIQRR61 pKa = 11.84NMLGMPSQRR70 pKa = 11.84LVSLRR75 pKa = 11.84QHH77 pKa = 6.28NSWTQRR83 pKa = 11.84MADD86 pKa = 3.72AGSGPAAGTVTMPLLANCPLSITSPLVSGWDD117 pKa = 3.44SDD119 pKa = 4.08PLFWEE124 pKa = 4.43MWSSVYY130 pKa = 10.66SHH132 pKa = 5.62YY133 pKa = 10.18TVVGVKK139 pKa = 8.25FTLKK143 pKa = 10.05TMWAAPTVTPTDD155 pKa = 3.1TEE157 pKa = 4.32GAVRR161 pKa = 11.84VPMRR165 pKa = 11.84VSLMVNTRR173 pKa = 11.84EE174 pKa = 4.3SIPDD178 pKa = 3.46YY179 pKa = 10.84VVPEE183 pKa = 3.9QLQEE187 pKa = 3.62QGLIHH192 pKa = 6.87WKK194 pKa = 10.7DD195 pKa = 3.56LMTSDD200 pKa = 4.62LSRR203 pKa = 11.84VHH205 pKa = 6.22VVEE208 pKa = 4.5KK209 pKa = 10.21FSAKK213 pKa = 10.16KK214 pKa = 10.22FFGLSSVSDD223 pKa = 4.13DD224 pKa = 3.91PSIGAPCTTYY234 pKa = 10.22PAALAWFLILVQKK247 pKa = 11.1VEE249 pKa = 4.15GTPFGGGGEE258 pKa = 4.21AGLPSSCSTVPSSFPSFSATRR279 pKa = 11.84SPRR282 pKa = 11.84PSRR285 pKa = 11.84RR286 pKa = 11.84PRR288 pKa = 11.84QANKK292 pKa = 10.58AIILSLFSCCLEE304 pKa = 4.23KK305 pKa = 10.96GAVYY309 pKa = 10.67YY310 pKa = 10.78GG311 pKa = 3.35
Molecular weight: 34.4 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
622 |
311 |
311 |
311.0 |
35.09 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.592 ± 0.117 | 3.376 ± 0.117 |
4.019 ± 0.819 | 5.305 ± 1.754 |
3.698 ± 0.117 | 7.235 ± 0.117 |
2.572 ± 0.234 | 3.055 ± 0.351 |
3.376 ± 0.117 | 9.486 ± 1.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.572 ± 0.702 | 3.055 ± 0.351 |
5.466 ± 0.936 | 3.698 ± 0.585 |
8.199 ± 0.351 | 7.717 ± 2.339 |
5.788 ± 0.468 | 8.682 ± 0.702 |
2.894 ± 0.234 | 3.215 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
