
Porcine associated porprismacovirus 4
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cremevirales; Smacoviridae; Porprismacovirus
Average proteome isoelectric point is 6.82
Get precalculated fractions of proteins
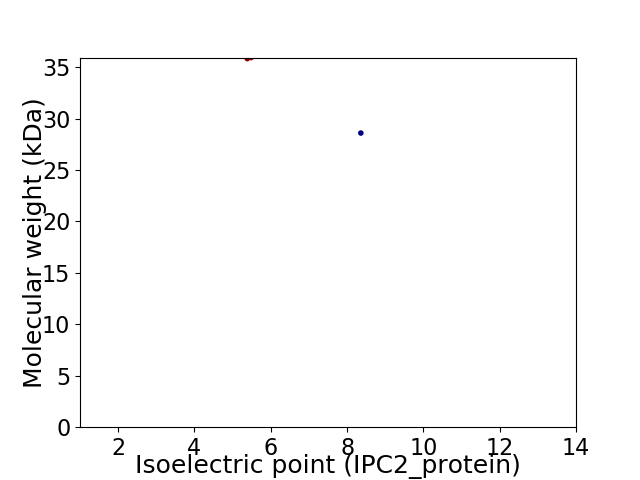
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
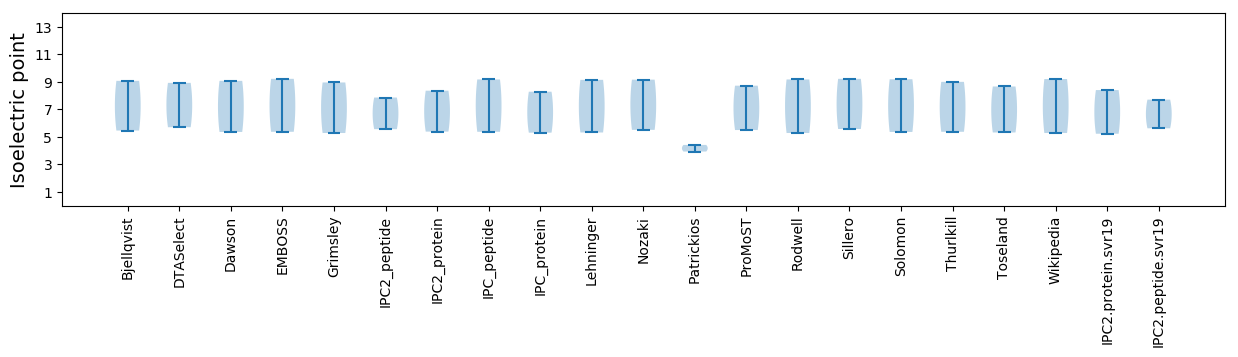
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A076VJK7|A0A076VJK7_9VIRU Rep protein OS=Porcine associated porprismacovirus 4 OX=2170120 PE=4 SV=1
MM1 pKa = 8.17VMVRR5 pKa = 11.84VSEE8 pKa = 4.34TYY10 pKa = 10.68DD11 pKa = 3.26LSTKK15 pKa = 8.72VGKK18 pKa = 9.42MGIVGIHH25 pKa = 4.95TPKK28 pKa = 10.68GNLITRR34 pKa = 11.84HH35 pKa = 5.39YY36 pKa = 10.55GGLIQNFKK44 pKa = 8.66YY45 pKa = 10.82AKK47 pKa = 9.94FVSCDD52 pKa = 3.15VALACASMLPADD64 pKa = 4.61PLQIGQEE71 pKa = 3.93AGAIAPQDD79 pKa = 3.54MFNPILYY86 pKa = 9.41RR87 pKa = 11.84AVSNEE92 pKa = 3.38SMNNFLAFLQSGAILNGAVDD112 pKa = 4.06KK113 pKa = 11.65NSVIDD118 pKa = 4.16LNNTSFTFNEE128 pKa = 4.12AEE130 pKa = 3.8VDD132 pKa = 3.77QFQLYY137 pKa = 10.4YY138 pKa = 11.11GLLSNSDD145 pKa = 3.32GWKK148 pKa = 10.17KK149 pKa = 11.0AMPQAGLEE157 pKa = 4.04MRR159 pKa = 11.84GLYY162 pKa = 9.98PLVFSVNVNTAMPSANGIGKK182 pKa = 8.88QFDD185 pKa = 3.82SEE187 pKa = 4.49FTGLSNTEE195 pKa = 3.63ASPAKK200 pKa = 10.35VGTVQWMRR208 pKa = 11.84GGAVRR213 pKa = 11.84MPRR216 pKa = 11.84FPTIRR221 pKa = 11.84FGTNEE226 pKa = 3.87QYY228 pKa = 11.73YY229 pKa = 10.67LIGDD233 pKa = 3.79VAGKK237 pKa = 10.4SDD239 pKa = 4.44LSDD242 pKa = 3.01MSMNAYY248 pKa = 9.86VAMIVLPPAKK258 pKa = 10.05LNQPYY263 pKa = 10.08YY264 pKa = 9.57RR265 pKa = 11.84LKK267 pKa = 9.62VTWTVEE273 pKa = 3.82FSEE276 pKa = 4.69IRR278 pKa = 11.84SQADD282 pKa = 2.9ILNWHH287 pKa = 6.44SLAAIGDD294 pKa = 4.01VSCGTDD300 pKa = 3.12YY301 pKa = 11.37AAQSKK306 pKa = 10.56AMARR310 pKa = 11.84TEE312 pKa = 4.3GMVDD316 pKa = 3.17TNGADD321 pKa = 3.38VTKK324 pKa = 11.01VMDD327 pKa = 5.29GII329 pKa = 4.31
MM1 pKa = 8.17VMVRR5 pKa = 11.84VSEE8 pKa = 4.34TYY10 pKa = 10.68DD11 pKa = 3.26LSTKK15 pKa = 8.72VGKK18 pKa = 9.42MGIVGIHH25 pKa = 4.95TPKK28 pKa = 10.68GNLITRR34 pKa = 11.84HH35 pKa = 5.39YY36 pKa = 10.55GGLIQNFKK44 pKa = 8.66YY45 pKa = 10.82AKK47 pKa = 9.94FVSCDD52 pKa = 3.15VALACASMLPADD64 pKa = 4.61PLQIGQEE71 pKa = 3.93AGAIAPQDD79 pKa = 3.54MFNPILYY86 pKa = 9.41RR87 pKa = 11.84AVSNEE92 pKa = 3.38SMNNFLAFLQSGAILNGAVDD112 pKa = 4.06KK113 pKa = 11.65NSVIDD118 pKa = 4.16LNNTSFTFNEE128 pKa = 4.12AEE130 pKa = 3.8VDD132 pKa = 3.77QFQLYY137 pKa = 10.4YY138 pKa = 11.11GLLSNSDD145 pKa = 3.32GWKK148 pKa = 10.17KK149 pKa = 11.0AMPQAGLEE157 pKa = 4.04MRR159 pKa = 11.84GLYY162 pKa = 9.98PLVFSVNVNTAMPSANGIGKK182 pKa = 8.88QFDD185 pKa = 3.82SEE187 pKa = 4.49FTGLSNTEE195 pKa = 3.63ASPAKK200 pKa = 10.35VGTVQWMRR208 pKa = 11.84GGAVRR213 pKa = 11.84MPRR216 pKa = 11.84FPTIRR221 pKa = 11.84FGTNEE226 pKa = 3.87QYY228 pKa = 11.73YY229 pKa = 10.67LIGDD233 pKa = 3.79VAGKK237 pKa = 10.4SDD239 pKa = 4.44LSDD242 pKa = 3.01MSMNAYY248 pKa = 9.86VAMIVLPPAKK258 pKa = 10.05LNQPYY263 pKa = 10.08YY264 pKa = 9.57RR265 pKa = 11.84LKK267 pKa = 9.62VTWTVEE273 pKa = 3.82FSEE276 pKa = 4.69IRR278 pKa = 11.84SQADD282 pKa = 2.9ILNWHH287 pKa = 6.44SLAAIGDD294 pKa = 4.01VSCGTDD300 pKa = 3.12YY301 pKa = 11.37AAQSKK306 pKa = 10.56AMARR310 pKa = 11.84TEE312 pKa = 4.3GMVDD316 pKa = 3.17TNGADD321 pKa = 3.38VTKK324 pKa = 11.01VMDD327 pKa = 5.29GII329 pKa = 4.31
Molecular weight: 35.82 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A076VJK7|A0A076VJK7_9VIRU Rep protein OS=Porcine associated porprismacovirus 4 OX=2170120 PE=4 SV=1
MM1 pKa = 6.19QTYY4 pKa = 10.75VMTIPRR10 pKa = 11.84TVSKK14 pKa = 10.35RR15 pKa = 11.84ALRR18 pKa = 11.84IMIEE22 pKa = 4.22KK23 pKa = 9.91NDD25 pKa = 3.67CKK27 pKa = 10.62KK28 pKa = 9.84WVIGKK33 pKa = 9.52EE34 pKa = 3.99EE35 pKa = 3.99GKK37 pKa = 10.26NGYY40 pKa = 8.24KK41 pKa = 9.71HH42 pKa = 4.18WQIRR46 pKa = 11.84IEE48 pKa = 4.08TSNDD52 pKa = 3.27DD53 pKa = 3.58FFEE56 pKa = 3.99WMQDD60 pKa = 3.7HH61 pKa = 7.07IPTAHH66 pKa = 6.05VEE68 pKa = 3.97KK69 pKa = 10.9SEE71 pKa = 4.2NGVDD75 pKa = 3.03ACRR78 pKa = 11.84YY79 pKa = 5.01EE80 pKa = 4.44TKK82 pKa = 10.22EE83 pKa = 3.92GQYY86 pKa = 11.43VMYY89 pKa = 10.2SDD91 pKa = 5.14RR92 pKa = 11.84VQNLMQRR99 pKa = 11.84YY100 pKa = 8.21GAFRR104 pKa = 11.84PNQKK108 pKa = 9.94RR109 pKa = 11.84AMQALQATNDD119 pKa = 3.61RR120 pKa = 11.84QVVVWYY126 pKa = 10.21DD127 pKa = 3.2EE128 pKa = 4.14TGNVGKK134 pKa = 10.24SWFTGALWEE143 pKa = 4.56RR144 pKa = 11.84GLAYY148 pKa = 7.93VTPPTIDD155 pKa = 3.16TVKK158 pKa = 11.09GLIQWVASCYY168 pKa = 9.78IDD170 pKa = 4.44GGWRR174 pKa = 11.84PYY176 pKa = 11.0VIIDD180 pKa = 4.19VPRR183 pKa = 11.84SWKK186 pKa = 9.71WSEE189 pKa = 3.8QLYY192 pKa = 10.74SAIEE196 pKa = 4.22SIKK199 pKa = 11.0DD200 pKa = 3.09GLIYY204 pKa = 9.1DD205 pKa = 4.01TRR207 pKa = 11.84YY208 pKa = 8.56HH209 pKa = 6.86SRR211 pKa = 11.84MINIRR216 pKa = 11.84GVKK219 pKa = 9.03VLVMTNTMPKK229 pKa = 9.86LDD231 pKa = 4.6KK232 pKa = 10.62LSKK235 pKa = 10.6DD236 pKa = 3.0RR237 pKa = 11.84WCICTFF243 pKa = 3.64
MM1 pKa = 6.19QTYY4 pKa = 10.75VMTIPRR10 pKa = 11.84TVSKK14 pKa = 10.35RR15 pKa = 11.84ALRR18 pKa = 11.84IMIEE22 pKa = 4.22KK23 pKa = 9.91NDD25 pKa = 3.67CKK27 pKa = 10.62KK28 pKa = 9.84WVIGKK33 pKa = 9.52EE34 pKa = 3.99EE35 pKa = 3.99GKK37 pKa = 10.26NGYY40 pKa = 8.24KK41 pKa = 9.71HH42 pKa = 4.18WQIRR46 pKa = 11.84IEE48 pKa = 4.08TSNDD52 pKa = 3.27DD53 pKa = 3.58FFEE56 pKa = 3.99WMQDD60 pKa = 3.7HH61 pKa = 7.07IPTAHH66 pKa = 6.05VEE68 pKa = 3.97KK69 pKa = 10.9SEE71 pKa = 4.2NGVDD75 pKa = 3.03ACRR78 pKa = 11.84YY79 pKa = 5.01EE80 pKa = 4.44TKK82 pKa = 10.22EE83 pKa = 3.92GQYY86 pKa = 11.43VMYY89 pKa = 10.2SDD91 pKa = 5.14RR92 pKa = 11.84VQNLMQRR99 pKa = 11.84YY100 pKa = 8.21GAFRR104 pKa = 11.84PNQKK108 pKa = 9.94RR109 pKa = 11.84AMQALQATNDD119 pKa = 3.61RR120 pKa = 11.84QVVVWYY126 pKa = 10.21DD127 pKa = 3.2EE128 pKa = 4.14TGNVGKK134 pKa = 10.24SWFTGALWEE143 pKa = 4.56RR144 pKa = 11.84GLAYY148 pKa = 7.93VTPPTIDD155 pKa = 3.16TVKK158 pKa = 11.09GLIQWVASCYY168 pKa = 9.78IDD170 pKa = 4.44GGWRR174 pKa = 11.84PYY176 pKa = 11.0VIIDD180 pKa = 4.19VPRR183 pKa = 11.84SWKK186 pKa = 9.71WSEE189 pKa = 3.8QLYY192 pKa = 10.74SAIEE196 pKa = 4.22SIKK199 pKa = 11.0DD200 pKa = 3.09GLIYY204 pKa = 9.1DD205 pKa = 4.01TRR207 pKa = 11.84YY208 pKa = 8.56HH209 pKa = 6.86SRR211 pKa = 11.84MINIRR216 pKa = 11.84GVKK219 pKa = 9.03VLVMTNTMPKK229 pKa = 9.86LDD231 pKa = 4.6KK232 pKa = 10.62LSKK235 pKa = 10.6DD236 pKa = 3.0RR237 pKa = 11.84WCICTFF243 pKa = 3.64
Molecular weight: 28.59 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
572 |
243 |
329 |
286.0 |
32.2 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.517 ± 1.974 | 1.399 ± 0.435 |
5.769 ± 0.266 | 4.371 ± 0.646 |
3.322 ± 0.835 | 7.517 ± 0.888 |
1.224 ± 0.279 | 6.119 ± 0.851 |
5.769 ± 1.081 | 6.294 ± 1.166 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
4.72 ± 0.399 | 5.42 ± 0.861 |
3.846 ± 0.366 | 4.545 ± 0.259 |
4.895 ± 1.387 | 6.294 ± 0.895 |
5.944 ± 0.423 | 7.867 ± 0.032 |
2.622 ± 1.257 | 4.545 ± 0.531 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
