
Belliella baltica (strain DSM 15883 / CIP 108006 / LMG 21964 / BA134)
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Cytophagia; Cytophagales; Cyclobacteriaceae; Belliella; Belliella baltica
Average proteome isoelectric point is 6.31
Get precalculated fractions of proteins
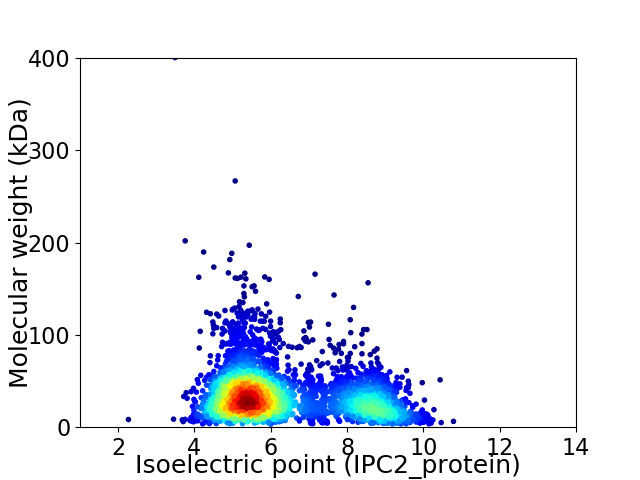
Virtual 2D-PAGE plot for 3609 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|I3Z5B7|I3Z5B7_BELBD Uncharacterized protein OS=Belliella baltica (strain DSM 15883 / CIP 108006 / LMG 21964 / BA134) OX=866536 GN=Belba_1846 PE=4 SV=1
MM1 pKa = 7.77KK2 pKa = 10.56SRR4 pKa = 11.84FKK6 pKa = 10.76SIKK9 pKa = 9.68FLLIFAGICVLSCIMPLNKK28 pKa = 10.07SFSQGFNNNEE38 pKa = 4.04WIFGYY43 pKa = 10.73CEE45 pKa = 4.96LDD47 pKa = 3.17TLKK50 pKa = 10.77RR51 pKa = 11.84IVSFGKK57 pKa = 10.52GEE59 pKa = 3.9EE60 pKa = 3.96PVIRR64 pKa = 11.84EE65 pKa = 3.53VSGAFNQNNSAVAVDD80 pKa = 4.78PINGNVVFYY89 pKa = 11.25SDD91 pKa = 4.14GEE93 pKa = 4.13LVYY96 pKa = 10.81NYY98 pKa = 10.14NNSPMQGALSINGNSSGRR116 pKa = 11.84QEE118 pKa = 3.71IGIGILEE125 pKa = 4.57FDD127 pKa = 4.01PNGDD131 pKa = 2.94KK132 pKa = 10.72LYY134 pKa = 10.84YY135 pKa = 9.83IFYY138 pKa = 8.72LTTGGQLQYY147 pKa = 11.48AVVDD151 pKa = 4.01MNSPGAAVGNEE162 pKa = 3.9PPQGRR167 pKa = 11.84VEE169 pKa = 4.1VFNRR173 pKa = 11.84SIGPATGSIAVVKK186 pKa = 10.05SAQSPSYY193 pKa = 10.41LISFNGGNLISRR205 pKa = 11.84RR206 pKa = 11.84IEE208 pKa = 3.71ATQGEE213 pKa = 4.84FTEE216 pKa = 4.5TDD218 pKa = 3.19DD219 pKa = 4.31VGIPFTPKK227 pKa = 10.45AIIFNEE233 pKa = 3.98ATGQLILIPEE243 pKa = 4.54NPNEE247 pKa = 4.86DD248 pKa = 3.18IVLVDD253 pKa = 4.13FDD255 pKa = 4.08TAEE258 pKa = 4.1GRR260 pKa = 11.84FGTVNTISQSGGVDD274 pKa = 3.71SIEE277 pKa = 4.18GAAFSPDD284 pKa = 2.76ADD286 pKa = 3.43YY287 pKa = 10.88IYY289 pKa = 10.71FSRR292 pKa = 11.84GNEE295 pKa = 4.14LFRR298 pKa = 11.84VPADD302 pKa = 4.58DD303 pKa = 5.74LDD305 pKa = 4.86ADD307 pKa = 4.18PEE309 pKa = 4.85LIPLEE314 pKa = 4.2NNVFRR319 pKa = 11.84IYY321 pKa = 10.32DD322 pKa = 3.54IKK324 pKa = 11.09VGPDD328 pKa = 2.64GRR330 pKa = 11.84LYY332 pKa = 10.79YY333 pKa = 9.94IYY335 pKa = 10.34EE336 pKa = 4.15EE337 pKa = 4.53VAGGPQLIGRR347 pKa = 11.84VDD349 pKa = 3.59NPDD352 pKa = 3.55EE353 pKa = 4.34EE354 pKa = 4.91DD355 pKa = 3.97LEE357 pKa = 4.32EE358 pKa = 4.82LEE360 pKa = 4.94KK361 pKa = 11.47DD362 pKa = 3.21EE363 pKa = 6.25DD364 pKa = 4.13PFDD367 pKa = 4.57GVDD370 pKa = 3.7FCGRR374 pKa = 11.84IFPQFAPNADD384 pKa = 3.33VEE386 pKa = 4.52ATVDD390 pKa = 4.25FTWEE394 pKa = 3.97PEE396 pKa = 4.45EE397 pKa = 4.35PCSNNPVQLTSVITPEE413 pKa = 3.59NYY415 pKa = 9.8RR416 pKa = 11.84PVSFEE421 pKa = 3.56WTFEE425 pKa = 4.13PEE427 pKa = 3.99LTDD430 pKa = 4.2EE431 pKa = 5.11DD432 pKa = 5.26GEE434 pKa = 4.47PLDD437 pKa = 3.56IDD439 pKa = 4.25YY440 pKa = 10.99NQEE443 pKa = 3.87HH444 pKa = 7.09LLLPAEE450 pKa = 4.44ATSGEE455 pKa = 4.77SVTATLTVTFADD467 pKa = 4.45GTTSTSDD474 pKa = 3.05PRR476 pKa = 11.84TINLSEE482 pKa = 4.37NNLEE486 pKa = 4.72ANFSPQDD493 pKa = 3.43TTVCEE498 pKa = 4.3SCFDD502 pKa = 4.08LMPLLEE508 pKa = 4.24VQQGEE513 pKa = 4.68GGQGGQGPGGPGGPGVGGPGGGNGVYY539 pKa = 10.21EE540 pKa = 4.37YY541 pKa = 10.66FWSNKK546 pKa = 8.34RR547 pKa = 11.84DD548 pKa = 3.77EE549 pKa = 4.64GWGPEE554 pKa = 4.14APNQICEE561 pKa = 4.21PGLYY565 pKa = 8.71WVLVKK570 pKa = 10.8EE571 pKa = 4.86PGSSCYY577 pKa = 10.14AYY579 pKa = 10.41ASIRR583 pKa = 11.84VRR585 pKa = 11.84MWDD588 pKa = 5.38LDD590 pKa = 3.92DD591 pKa = 3.55QSNNIWYY598 pKa = 9.33FGDD601 pKa = 3.68GAGIDD606 pKa = 4.15FNEE609 pKa = 4.8DD610 pKa = 3.09PAGILPTPRR619 pKa = 11.84PVDD622 pKa = 3.2PRR624 pKa = 11.84HH625 pKa = 5.89PQNIPAGTTTISDD638 pKa = 3.26EE639 pKa = 4.2TGQVLFYY646 pKa = 10.75TDD648 pKa = 4.92GSTVWDD654 pKa = 4.2LYY656 pKa = 10.74GDD658 pKa = 3.86VMADD662 pKa = 3.45GEE664 pKa = 4.98DD665 pKa = 3.22IGGSNQASEE674 pKa = 4.55GVIAVAIPQEE684 pKa = 3.96EE685 pKa = 4.82TIFYY689 pKa = 10.81LFTTSTAADD698 pKa = 3.66GNSQVKK704 pKa = 10.12FSLVDD709 pKa = 3.59IKK711 pKa = 11.36AEE713 pKa = 3.95NPTGVGNVVSKK724 pKa = 11.44DD725 pKa = 3.26NFLFSPATQQSAAIAAGDD743 pKa = 3.89TTWVLFHH750 pKa = 7.06EE751 pKa = 5.11LGNNTFRR758 pKa = 11.84AYY760 pKa = 9.72PVGSLGIGPPVFSSVGSTHH779 pKa = 7.13GYY781 pKa = 7.13DD782 pKa = 3.11TGVGSMKK789 pKa = 10.4FSPDD793 pKa = 2.97GTKK796 pKa = 10.45VAVTIQDD803 pKa = 4.02GACSRR808 pKa = 11.84VEE810 pKa = 5.24LFDD813 pKa = 4.78FDD815 pKa = 3.74QSTGRR820 pKa = 11.84LTEE823 pKa = 4.05YY824 pKa = 11.09ASVDD828 pKa = 3.98LDD830 pKa = 4.33CNDD833 pKa = 3.78DD834 pKa = 3.63DD835 pKa = 5.04VYY837 pKa = 11.54GVEE840 pKa = 5.07FSNDD844 pKa = 2.74SDD846 pKa = 4.54RR847 pKa = 11.84IFVTYY852 pKa = 9.85TGDD855 pKa = 3.35GGKK858 pKa = 9.59VEE860 pKa = 4.18EE861 pKa = 5.68FIIQSPSSQDD871 pKa = 3.1EE872 pKa = 4.38DD873 pKa = 5.61DD874 pKa = 5.28DD875 pKa = 5.06LSCGACFEE883 pKa = 4.57RR884 pKa = 11.84ATTSAQRR891 pKa = 11.84AQCILDD897 pKa = 3.3SRR899 pKa = 11.84TDD901 pKa = 3.41LRR903 pKa = 11.84INGPIGAIQMGPMGDD918 pKa = 2.92IYY920 pKa = 10.72VARR923 pKa = 11.84PGQNFLGTIQPGGLCEE939 pKa = 4.73LSTGTTEE946 pKa = 5.45GFQLLPGTTSNLGLPSFVQQSGSSIPEE973 pKa = 3.86PAISGPDD980 pKa = 3.6RR981 pKa = 11.84LCLDD985 pKa = 4.17PEE987 pKa = 4.44NGAEE991 pKa = 4.23GLFEE995 pKa = 4.43GAGEE999 pKa = 4.14PDD1001 pKa = 2.98IDD1003 pKa = 4.08SYY1005 pKa = 11.62FWTIVHH1011 pKa = 6.89EE1012 pKa = 4.72DD1013 pKa = 3.97GEE1015 pKa = 4.76VIEE1018 pKa = 5.2SDD1020 pKa = 2.98LGGPGEE1026 pKa = 4.32EE1027 pKa = 4.39NQTLTYY1033 pKa = 10.01IFQRR1037 pKa = 11.84EE1038 pKa = 3.89GLYY1041 pKa = 9.99TVTLRR1046 pKa = 11.84VDD1048 pKa = 3.04RR1049 pKa = 11.84CGDD1052 pKa = 3.38PEE1054 pKa = 4.72HH1055 pKa = 6.74YY1056 pKa = 10.41NKK1058 pKa = 10.48NNSIEE1063 pKa = 4.26VLVVAPPEE1071 pKa = 3.84ITLEE1075 pKa = 4.02NDD1077 pKa = 2.84VTLCSGTPVTLTAIDD1092 pKa = 4.97GYY1094 pKa = 11.44DD1095 pKa = 3.76PADD1098 pKa = 3.52GLYY1101 pKa = 10.83DD1102 pKa = 3.85FEE1104 pKa = 4.45WRR1106 pKa = 11.84NAAGIQFGDD1115 pKa = 3.78EE1116 pKa = 4.03NSNEE1120 pKa = 3.86ITVKK1124 pKa = 10.52EE1125 pKa = 3.86EE1126 pKa = 3.72SIYY1129 pKa = 10.62TVTVSYY1135 pKa = 11.19RR1136 pKa = 11.84NTNQDD1141 pKa = 3.08EE1142 pKa = 4.69EE1143 pKa = 5.05FFDD1146 pKa = 3.95ACPATRR1152 pKa = 11.84SVFVGPAFEE1161 pKa = 5.4FDD1163 pKa = 3.28LTQTAEE1169 pKa = 4.26EE1170 pKa = 4.33VCYY1173 pKa = 11.03DD1174 pKa = 3.35EE1175 pKa = 4.53TLVVFAPDD1183 pKa = 3.78TPVSGEE1189 pKa = 3.48WFYY1192 pKa = 11.72QRR1194 pKa = 11.84VGDD1197 pKa = 4.07PNRR1200 pKa = 11.84VQFPVGLAFEE1210 pKa = 5.29LEE1212 pKa = 4.14ITPSIDD1218 pKa = 3.29LPGPGLYY1225 pKa = 10.08QIIFVTEE1232 pKa = 3.91DD1233 pKa = 4.36PIVEE1237 pKa = 4.27GCLVEE1242 pKa = 5.19KK1243 pKa = 10.81VLEE1246 pKa = 4.27LEE1248 pKa = 4.5VFPLPNFEE1256 pKa = 5.47IVILTNAEE1264 pKa = 4.07DD1265 pKa = 4.12CNSPEE1270 pKa = 3.82GSFEE1274 pKa = 3.93IEE1276 pKa = 3.64ALIAMSVLEE1285 pKa = 4.02IMEE1288 pKa = 4.57TGDD1291 pKa = 3.17VFQNVSAGDD1300 pKa = 4.0FFTLTDD1306 pKa = 4.71LEE1308 pKa = 4.66PGNYY1312 pKa = 7.18TVRR1315 pKa = 11.84GQTDD1319 pKa = 4.18FGCEE1323 pKa = 3.92FTRR1326 pKa = 11.84TATIQNVNPPQGFEE1340 pKa = 4.4NITLDD1345 pKa = 4.43STDD1348 pKa = 3.42EE1349 pKa = 4.19SCGVNEE1355 pKa = 5.39IIPGSITIEE1364 pKa = 3.73LSAAVQGTYY1373 pKa = 10.3TITRR1377 pKa = 11.84QGDD1380 pKa = 3.73GQEE1383 pKa = 4.05FTGALDD1389 pKa = 3.72GQTTVIEE1396 pKa = 4.58IGHH1399 pKa = 6.84GDD1401 pKa = 3.66YY1402 pKa = 10.97AVQVEE1407 pKa = 4.65DD1408 pKa = 5.02ANGCAIPVGDD1418 pKa = 3.54VTVARR1423 pKa = 11.84RR1424 pKa = 11.84FLADD1428 pKa = 4.04FNVPSNIVACEE1439 pKa = 3.93SFEE1442 pKa = 5.36LEE1444 pKa = 4.92ISTQQNLVFTVTDD1457 pKa = 4.07PNGDD1461 pKa = 4.77LINADD1466 pKa = 3.09ADD1468 pKa = 4.45GIYY1471 pKa = 10.35LLNISGVYY1479 pKa = 8.8TVFGEE1484 pKa = 4.51DD1485 pKa = 3.95PDD1487 pKa = 5.29GVACPRR1493 pKa = 11.84EE1494 pKa = 3.85RR1495 pKa = 11.84TINLTINDD1503 pKa = 5.27PIQYY1507 pKa = 9.24EE1508 pKa = 4.24LSAPQVDD1515 pKa = 4.55CVDD1518 pKa = 3.59GVSYY1522 pKa = 10.27EE1523 pKa = 4.42AIILDD1528 pKa = 3.92GTDD1531 pKa = 3.34PASVFFFWRR1540 pKa = 11.84NEE1542 pKa = 3.57SSQVVGRR1549 pKa = 11.84SQRR1552 pKa = 11.84FFPSAPGQYY1561 pKa = 9.37TLDD1564 pKa = 3.44VQPRR1568 pKa = 11.84VGSNCPTPPIPFEE1581 pKa = 4.18VEE1583 pKa = 3.65EE1584 pKa = 4.15FAAAVDD1590 pKa = 4.8FEE1592 pKa = 5.93LEE1594 pKa = 4.16VQPFCVGEE1602 pKa = 3.76PSTFVNLLIDD1612 pKa = 4.74ADD1614 pKa = 4.19DD1615 pKa = 3.97MDD1617 pKa = 4.7AVGQIRR1623 pKa = 11.84WFFVQGGNRR1632 pKa = 11.84TPLPANDD1639 pKa = 4.3DD1640 pKa = 3.58ATMVSVDD1647 pKa = 3.46QANQGTYY1654 pKa = 7.75QVEE1657 pKa = 4.58VYY1659 pKa = 11.25NNFGCLVGRR1668 pKa = 11.84DD1669 pKa = 3.97QIQVIQSTLVAPVLQATYY1687 pKa = 10.05IICALEE1693 pKa = 4.27NIVFQLDD1700 pKa = 3.03PGQYY1704 pKa = 10.93DD1705 pKa = 3.22NYY1707 pKa = 9.69EE1708 pKa = 3.96WYY1710 pKa = 10.9LEE1712 pKa = 4.52GEE1714 pKa = 4.68DD1715 pKa = 4.69EE1716 pKa = 5.6PISTDD1721 pKa = 4.51PIFTPQQEE1729 pKa = 4.33GNYY1732 pKa = 9.02RR1733 pKa = 11.84LVVFDD1738 pKa = 4.51EE1739 pKa = 4.61LGCVEE1744 pKa = 5.11EE1745 pKa = 4.22ITFEE1749 pKa = 4.17VVEE1752 pKa = 4.3DD1753 pKa = 3.76CEE1755 pKa = 4.62LRR1757 pKa = 11.84VRR1759 pKa = 11.84FPNAMSPSDD1768 pKa = 3.52ASKK1771 pKa = 11.0QFLIYY1776 pKa = 10.53TNDD1779 pKa = 4.02FIDD1782 pKa = 3.83EE1783 pKa = 4.33LSVYY1787 pKa = 10.0IYY1789 pKa = 10.73NRR1791 pKa = 11.84WGEE1794 pKa = 4.18LIFYY1798 pKa = 9.63CEE1800 pKa = 3.96QSNLPGDD1807 pKa = 3.9AVGGYY1812 pKa = 9.14CPWDD1816 pKa = 3.36GTVNGQKK1823 pKa = 10.09VPIGTYY1829 pKa = 8.39PVVVRR1834 pKa = 11.84FRR1836 pKa = 11.84SNNQNITKK1844 pKa = 8.0TLRR1847 pKa = 11.84EE1848 pKa = 4.4AIVVIEE1854 pKa = 3.99
MM1 pKa = 7.77KK2 pKa = 10.56SRR4 pKa = 11.84FKK6 pKa = 10.76SIKK9 pKa = 9.68FLLIFAGICVLSCIMPLNKK28 pKa = 10.07SFSQGFNNNEE38 pKa = 4.04WIFGYY43 pKa = 10.73CEE45 pKa = 4.96LDD47 pKa = 3.17TLKK50 pKa = 10.77RR51 pKa = 11.84IVSFGKK57 pKa = 10.52GEE59 pKa = 3.9EE60 pKa = 3.96PVIRR64 pKa = 11.84EE65 pKa = 3.53VSGAFNQNNSAVAVDD80 pKa = 4.78PINGNVVFYY89 pKa = 11.25SDD91 pKa = 4.14GEE93 pKa = 4.13LVYY96 pKa = 10.81NYY98 pKa = 10.14NNSPMQGALSINGNSSGRR116 pKa = 11.84QEE118 pKa = 3.71IGIGILEE125 pKa = 4.57FDD127 pKa = 4.01PNGDD131 pKa = 2.94KK132 pKa = 10.72LYY134 pKa = 10.84YY135 pKa = 9.83IFYY138 pKa = 8.72LTTGGQLQYY147 pKa = 11.48AVVDD151 pKa = 4.01MNSPGAAVGNEE162 pKa = 3.9PPQGRR167 pKa = 11.84VEE169 pKa = 4.1VFNRR173 pKa = 11.84SIGPATGSIAVVKK186 pKa = 10.05SAQSPSYY193 pKa = 10.41LISFNGGNLISRR205 pKa = 11.84RR206 pKa = 11.84IEE208 pKa = 3.71ATQGEE213 pKa = 4.84FTEE216 pKa = 4.5TDD218 pKa = 3.19DD219 pKa = 4.31VGIPFTPKK227 pKa = 10.45AIIFNEE233 pKa = 3.98ATGQLILIPEE243 pKa = 4.54NPNEE247 pKa = 4.86DD248 pKa = 3.18IVLVDD253 pKa = 4.13FDD255 pKa = 4.08TAEE258 pKa = 4.1GRR260 pKa = 11.84FGTVNTISQSGGVDD274 pKa = 3.71SIEE277 pKa = 4.18GAAFSPDD284 pKa = 2.76ADD286 pKa = 3.43YY287 pKa = 10.88IYY289 pKa = 10.71FSRR292 pKa = 11.84GNEE295 pKa = 4.14LFRR298 pKa = 11.84VPADD302 pKa = 4.58DD303 pKa = 5.74LDD305 pKa = 4.86ADD307 pKa = 4.18PEE309 pKa = 4.85LIPLEE314 pKa = 4.2NNVFRR319 pKa = 11.84IYY321 pKa = 10.32DD322 pKa = 3.54IKK324 pKa = 11.09VGPDD328 pKa = 2.64GRR330 pKa = 11.84LYY332 pKa = 10.79YY333 pKa = 9.94IYY335 pKa = 10.34EE336 pKa = 4.15EE337 pKa = 4.53VAGGPQLIGRR347 pKa = 11.84VDD349 pKa = 3.59NPDD352 pKa = 3.55EE353 pKa = 4.34EE354 pKa = 4.91DD355 pKa = 3.97LEE357 pKa = 4.32EE358 pKa = 4.82LEE360 pKa = 4.94KK361 pKa = 11.47DD362 pKa = 3.21EE363 pKa = 6.25DD364 pKa = 4.13PFDD367 pKa = 4.57GVDD370 pKa = 3.7FCGRR374 pKa = 11.84IFPQFAPNADD384 pKa = 3.33VEE386 pKa = 4.52ATVDD390 pKa = 4.25FTWEE394 pKa = 3.97PEE396 pKa = 4.45EE397 pKa = 4.35PCSNNPVQLTSVITPEE413 pKa = 3.59NYY415 pKa = 9.8RR416 pKa = 11.84PVSFEE421 pKa = 3.56WTFEE425 pKa = 4.13PEE427 pKa = 3.99LTDD430 pKa = 4.2EE431 pKa = 5.11DD432 pKa = 5.26GEE434 pKa = 4.47PLDD437 pKa = 3.56IDD439 pKa = 4.25YY440 pKa = 10.99NQEE443 pKa = 3.87HH444 pKa = 7.09LLLPAEE450 pKa = 4.44ATSGEE455 pKa = 4.77SVTATLTVTFADD467 pKa = 4.45GTTSTSDD474 pKa = 3.05PRR476 pKa = 11.84TINLSEE482 pKa = 4.37NNLEE486 pKa = 4.72ANFSPQDD493 pKa = 3.43TTVCEE498 pKa = 4.3SCFDD502 pKa = 4.08LMPLLEE508 pKa = 4.24VQQGEE513 pKa = 4.68GGQGGQGPGGPGGPGVGGPGGGNGVYY539 pKa = 10.21EE540 pKa = 4.37YY541 pKa = 10.66FWSNKK546 pKa = 8.34RR547 pKa = 11.84DD548 pKa = 3.77EE549 pKa = 4.64GWGPEE554 pKa = 4.14APNQICEE561 pKa = 4.21PGLYY565 pKa = 8.71WVLVKK570 pKa = 10.8EE571 pKa = 4.86PGSSCYY577 pKa = 10.14AYY579 pKa = 10.41ASIRR583 pKa = 11.84VRR585 pKa = 11.84MWDD588 pKa = 5.38LDD590 pKa = 3.92DD591 pKa = 3.55QSNNIWYY598 pKa = 9.33FGDD601 pKa = 3.68GAGIDD606 pKa = 4.15FNEE609 pKa = 4.8DD610 pKa = 3.09PAGILPTPRR619 pKa = 11.84PVDD622 pKa = 3.2PRR624 pKa = 11.84HH625 pKa = 5.89PQNIPAGTTTISDD638 pKa = 3.26EE639 pKa = 4.2TGQVLFYY646 pKa = 10.75TDD648 pKa = 4.92GSTVWDD654 pKa = 4.2LYY656 pKa = 10.74GDD658 pKa = 3.86VMADD662 pKa = 3.45GEE664 pKa = 4.98DD665 pKa = 3.22IGGSNQASEE674 pKa = 4.55GVIAVAIPQEE684 pKa = 3.96EE685 pKa = 4.82TIFYY689 pKa = 10.81LFTTSTAADD698 pKa = 3.66GNSQVKK704 pKa = 10.12FSLVDD709 pKa = 3.59IKK711 pKa = 11.36AEE713 pKa = 3.95NPTGVGNVVSKK724 pKa = 11.44DD725 pKa = 3.26NFLFSPATQQSAAIAAGDD743 pKa = 3.89TTWVLFHH750 pKa = 7.06EE751 pKa = 5.11LGNNTFRR758 pKa = 11.84AYY760 pKa = 9.72PVGSLGIGPPVFSSVGSTHH779 pKa = 7.13GYY781 pKa = 7.13DD782 pKa = 3.11TGVGSMKK789 pKa = 10.4FSPDD793 pKa = 2.97GTKK796 pKa = 10.45VAVTIQDD803 pKa = 4.02GACSRR808 pKa = 11.84VEE810 pKa = 5.24LFDD813 pKa = 4.78FDD815 pKa = 3.74QSTGRR820 pKa = 11.84LTEE823 pKa = 4.05YY824 pKa = 11.09ASVDD828 pKa = 3.98LDD830 pKa = 4.33CNDD833 pKa = 3.78DD834 pKa = 3.63DD835 pKa = 5.04VYY837 pKa = 11.54GVEE840 pKa = 5.07FSNDD844 pKa = 2.74SDD846 pKa = 4.54RR847 pKa = 11.84IFVTYY852 pKa = 9.85TGDD855 pKa = 3.35GGKK858 pKa = 9.59VEE860 pKa = 4.18EE861 pKa = 5.68FIIQSPSSQDD871 pKa = 3.1EE872 pKa = 4.38DD873 pKa = 5.61DD874 pKa = 5.28DD875 pKa = 5.06LSCGACFEE883 pKa = 4.57RR884 pKa = 11.84ATTSAQRR891 pKa = 11.84AQCILDD897 pKa = 3.3SRR899 pKa = 11.84TDD901 pKa = 3.41LRR903 pKa = 11.84INGPIGAIQMGPMGDD918 pKa = 2.92IYY920 pKa = 10.72VARR923 pKa = 11.84PGQNFLGTIQPGGLCEE939 pKa = 4.73LSTGTTEE946 pKa = 5.45GFQLLPGTTSNLGLPSFVQQSGSSIPEE973 pKa = 3.86PAISGPDD980 pKa = 3.6RR981 pKa = 11.84LCLDD985 pKa = 4.17PEE987 pKa = 4.44NGAEE991 pKa = 4.23GLFEE995 pKa = 4.43GAGEE999 pKa = 4.14PDD1001 pKa = 2.98IDD1003 pKa = 4.08SYY1005 pKa = 11.62FWTIVHH1011 pKa = 6.89EE1012 pKa = 4.72DD1013 pKa = 3.97GEE1015 pKa = 4.76VIEE1018 pKa = 5.2SDD1020 pKa = 2.98LGGPGEE1026 pKa = 4.32EE1027 pKa = 4.39NQTLTYY1033 pKa = 10.01IFQRR1037 pKa = 11.84EE1038 pKa = 3.89GLYY1041 pKa = 9.99TVTLRR1046 pKa = 11.84VDD1048 pKa = 3.04RR1049 pKa = 11.84CGDD1052 pKa = 3.38PEE1054 pKa = 4.72HH1055 pKa = 6.74YY1056 pKa = 10.41NKK1058 pKa = 10.48NNSIEE1063 pKa = 4.26VLVVAPPEE1071 pKa = 3.84ITLEE1075 pKa = 4.02NDD1077 pKa = 2.84VTLCSGTPVTLTAIDD1092 pKa = 4.97GYY1094 pKa = 11.44DD1095 pKa = 3.76PADD1098 pKa = 3.52GLYY1101 pKa = 10.83DD1102 pKa = 3.85FEE1104 pKa = 4.45WRR1106 pKa = 11.84NAAGIQFGDD1115 pKa = 3.78EE1116 pKa = 4.03NSNEE1120 pKa = 3.86ITVKK1124 pKa = 10.52EE1125 pKa = 3.86EE1126 pKa = 3.72SIYY1129 pKa = 10.62TVTVSYY1135 pKa = 11.19RR1136 pKa = 11.84NTNQDD1141 pKa = 3.08EE1142 pKa = 4.69EE1143 pKa = 5.05FFDD1146 pKa = 3.95ACPATRR1152 pKa = 11.84SVFVGPAFEE1161 pKa = 5.4FDD1163 pKa = 3.28LTQTAEE1169 pKa = 4.26EE1170 pKa = 4.33VCYY1173 pKa = 11.03DD1174 pKa = 3.35EE1175 pKa = 4.53TLVVFAPDD1183 pKa = 3.78TPVSGEE1189 pKa = 3.48WFYY1192 pKa = 11.72QRR1194 pKa = 11.84VGDD1197 pKa = 4.07PNRR1200 pKa = 11.84VQFPVGLAFEE1210 pKa = 5.29LEE1212 pKa = 4.14ITPSIDD1218 pKa = 3.29LPGPGLYY1225 pKa = 10.08QIIFVTEE1232 pKa = 3.91DD1233 pKa = 4.36PIVEE1237 pKa = 4.27GCLVEE1242 pKa = 5.19KK1243 pKa = 10.81VLEE1246 pKa = 4.27LEE1248 pKa = 4.5VFPLPNFEE1256 pKa = 5.47IVILTNAEE1264 pKa = 4.07DD1265 pKa = 4.12CNSPEE1270 pKa = 3.82GSFEE1274 pKa = 3.93IEE1276 pKa = 3.64ALIAMSVLEE1285 pKa = 4.02IMEE1288 pKa = 4.57TGDD1291 pKa = 3.17VFQNVSAGDD1300 pKa = 4.0FFTLTDD1306 pKa = 4.71LEE1308 pKa = 4.66PGNYY1312 pKa = 7.18TVRR1315 pKa = 11.84GQTDD1319 pKa = 4.18FGCEE1323 pKa = 3.92FTRR1326 pKa = 11.84TATIQNVNPPQGFEE1340 pKa = 4.4NITLDD1345 pKa = 4.43STDD1348 pKa = 3.42EE1349 pKa = 4.19SCGVNEE1355 pKa = 5.39IIPGSITIEE1364 pKa = 3.73LSAAVQGTYY1373 pKa = 10.3TITRR1377 pKa = 11.84QGDD1380 pKa = 3.73GQEE1383 pKa = 4.05FTGALDD1389 pKa = 3.72GQTTVIEE1396 pKa = 4.58IGHH1399 pKa = 6.84GDD1401 pKa = 3.66YY1402 pKa = 10.97AVQVEE1407 pKa = 4.65DD1408 pKa = 5.02ANGCAIPVGDD1418 pKa = 3.54VTVARR1423 pKa = 11.84RR1424 pKa = 11.84FLADD1428 pKa = 4.04FNVPSNIVACEE1439 pKa = 3.93SFEE1442 pKa = 5.36LEE1444 pKa = 4.92ISTQQNLVFTVTDD1457 pKa = 4.07PNGDD1461 pKa = 4.77LINADD1466 pKa = 3.09ADD1468 pKa = 4.45GIYY1471 pKa = 10.35LLNISGVYY1479 pKa = 8.8TVFGEE1484 pKa = 4.51DD1485 pKa = 3.95PDD1487 pKa = 5.29GVACPRR1493 pKa = 11.84EE1494 pKa = 3.85RR1495 pKa = 11.84TINLTINDD1503 pKa = 5.27PIQYY1507 pKa = 9.24EE1508 pKa = 4.24LSAPQVDD1515 pKa = 4.55CVDD1518 pKa = 3.59GVSYY1522 pKa = 10.27EE1523 pKa = 4.42AIILDD1528 pKa = 3.92GTDD1531 pKa = 3.34PASVFFFWRR1540 pKa = 11.84NEE1542 pKa = 3.57SSQVVGRR1549 pKa = 11.84SQRR1552 pKa = 11.84FFPSAPGQYY1561 pKa = 9.37TLDD1564 pKa = 3.44VQPRR1568 pKa = 11.84VGSNCPTPPIPFEE1581 pKa = 4.18VEE1583 pKa = 3.65EE1584 pKa = 4.15FAAAVDD1590 pKa = 4.8FEE1592 pKa = 5.93LEE1594 pKa = 4.16VQPFCVGEE1602 pKa = 3.76PSTFVNLLIDD1612 pKa = 4.74ADD1614 pKa = 4.19DD1615 pKa = 3.97MDD1617 pKa = 4.7AVGQIRR1623 pKa = 11.84WFFVQGGNRR1632 pKa = 11.84TPLPANDD1639 pKa = 4.3DD1640 pKa = 3.58ATMVSVDD1647 pKa = 3.46QANQGTYY1654 pKa = 7.75QVEE1657 pKa = 4.58VYY1659 pKa = 11.25NNFGCLVGRR1668 pKa = 11.84DD1669 pKa = 3.97QIQVIQSTLVAPVLQATYY1687 pKa = 10.05IICALEE1693 pKa = 4.27NIVFQLDD1700 pKa = 3.03PGQYY1704 pKa = 10.93DD1705 pKa = 3.22NYY1707 pKa = 9.69EE1708 pKa = 3.96WYY1710 pKa = 10.9LEE1712 pKa = 4.52GEE1714 pKa = 4.68DD1715 pKa = 4.69EE1716 pKa = 5.6PISTDD1721 pKa = 4.51PIFTPQQEE1729 pKa = 4.33GNYY1732 pKa = 9.02RR1733 pKa = 11.84LVVFDD1738 pKa = 4.51EE1739 pKa = 4.61LGCVEE1744 pKa = 5.11EE1745 pKa = 4.22ITFEE1749 pKa = 4.17VVEE1752 pKa = 4.3DD1753 pKa = 3.76CEE1755 pKa = 4.62LRR1757 pKa = 11.84VRR1759 pKa = 11.84FPNAMSPSDD1768 pKa = 3.52ASKK1771 pKa = 11.0QFLIYY1776 pKa = 10.53TNDD1779 pKa = 4.02FIDD1782 pKa = 3.83EE1783 pKa = 4.33LSVYY1787 pKa = 10.0IYY1789 pKa = 10.73NRR1791 pKa = 11.84WGEE1794 pKa = 4.18LIFYY1798 pKa = 9.63CEE1800 pKa = 3.96QSNLPGDD1807 pKa = 3.9AVGGYY1812 pKa = 9.14CPWDD1816 pKa = 3.36GTVNGQKK1823 pKa = 10.09VPIGTYY1829 pKa = 8.39PVVVRR1834 pKa = 11.84FRR1836 pKa = 11.84SNNQNITKK1844 pKa = 8.0TLRR1847 pKa = 11.84EE1848 pKa = 4.4AIVVIEE1854 pKa = 3.99
Molecular weight: 202.01 kDa
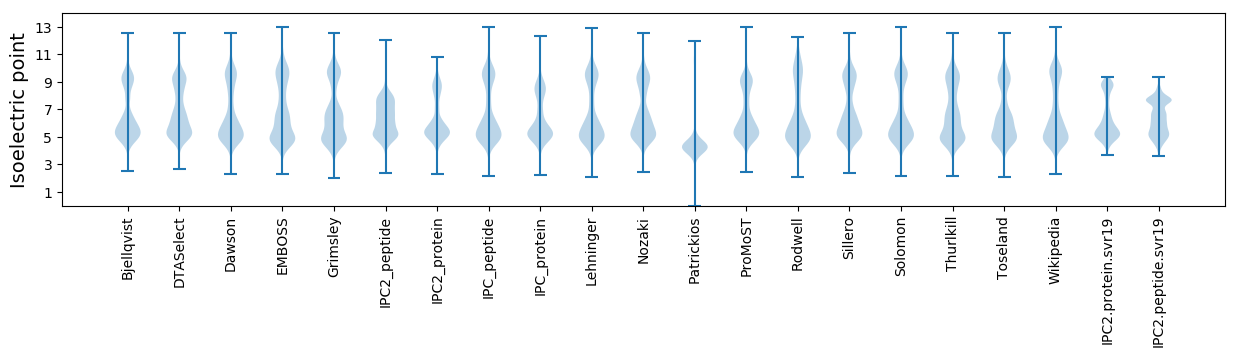
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|I3Z591|I3Z591_BELBD PIF1 helicase/Helicase OS=Belliella baltica (strain DSM 15883 / CIP 108006 / LMG 21964 / BA134) OX=866536 GN=Belba_1817 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84RR10 pKa = 11.84KK11 pKa = 9.62RR12 pKa = 11.84RR13 pKa = 11.84NKK15 pKa = 9.5HH16 pKa = 3.99GFRR19 pKa = 11.84EE20 pKa = 4.18RR21 pKa = 11.84MSTANGRR28 pKa = 11.84RR29 pKa = 11.84VIRR32 pKa = 11.84SRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.0GRR39 pKa = 11.84HH40 pKa = 5.41KK41 pKa = 10.43LTVSSEE47 pKa = 4.01KK48 pKa = 9.88TLKK51 pKa = 10.51KK52 pKa = 10.72
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84RR10 pKa = 11.84KK11 pKa = 9.62RR12 pKa = 11.84RR13 pKa = 11.84NKK15 pKa = 9.5HH16 pKa = 3.99GFRR19 pKa = 11.84EE20 pKa = 4.18RR21 pKa = 11.84MSTANGRR28 pKa = 11.84RR29 pKa = 11.84VIRR32 pKa = 11.84SRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.0GRR39 pKa = 11.84HH40 pKa = 5.41KK41 pKa = 10.43LTVSSEE47 pKa = 4.01KK48 pKa = 9.88TLKK51 pKa = 10.51KK52 pKa = 10.72
Molecular weight: 6.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1180055 |
29 |
3807 |
327.0 |
36.98 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.334 ± 0.039 | 0.678 ± 0.013 |
5.461 ± 0.031 | 7.187 ± 0.039 |
5.459 ± 0.034 | 6.636 ± 0.04 |
1.779 ± 0.025 | 7.868 ± 0.038 |
7.427 ± 0.05 | 9.706 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.405 ± 0.019 | 5.482 ± 0.035 |
3.594 ± 0.024 | 3.621 ± 0.02 |
3.899 ± 0.03 | 6.564 ± 0.029 |
4.951 ± 0.039 | 6.049 ± 0.04 |
1.12 ± 0.015 | 3.782 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
