
Simian T-lymphotropic virus 1
Taxonomy: Viruses; Riboviria; Pararnavirae; Artverviricota; Revtraviricetes; Ortervirales; Retroviridae; Orthoretrovirinae; Deltaretrovirus; Primate T-lymphotropic virus 1
Average proteome isoelectric point is 7.0
Get precalculated fractions of proteins
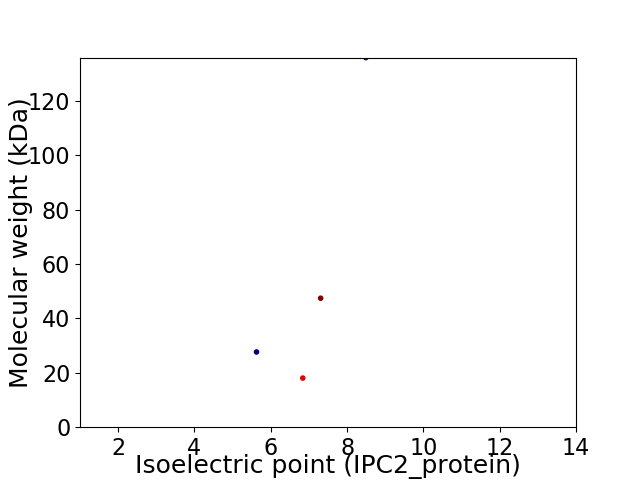
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q9WS53|Q9WS53_9STL1 Env polyprotein OS=Simian T-lymphotropic virus 1 OX=33747 PE=4 SV=1
MM1 pKa = 7.74RR2 pKa = 11.84KK3 pKa = 9.61YY4 pKa = 10.63SPFRR8 pKa = 11.84NGYY11 pKa = 7.91MEE13 pKa = 4.35PTLGQHH19 pKa = 6.94LPTLSFPDD27 pKa = 3.77PGLRR31 pKa = 11.84PQNLYY36 pKa = 8.25TLWGDD41 pKa = 4.07SVVCLYY47 pKa = 10.65LYY49 pKa = 10.33QLSPPITWPLPPHH62 pKa = 7.02VIFCHH67 pKa = 6.21PGQLGAFLTNVPYY80 pKa = 10.94KK81 pKa = 10.66RR82 pKa = 11.84MEE84 pKa = 3.93EE85 pKa = 3.84LLYY88 pKa = 10.66KK89 pKa = 10.55ISLTTGALIILPEE102 pKa = 4.78DD103 pKa = 3.97CLPTTLFQPARR114 pKa = 11.84APVTLTAWQNGLLPFHH130 pKa = 6.75STLTTPGLIWTFTDD144 pKa = 3.88GTPMVSGPCPKK155 pKa = 10.26DD156 pKa = 3.49GQPSLVLQSSSFIFHH171 pKa = 7.35KK172 pKa = 10.59FQTKK176 pKa = 9.83AYY178 pKa = 8.81HH179 pKa = 6.37PSFLLSHH186 pKa = 6.6GLIQYY191 pKa = 10.76SSFHH195 pKa = 5.98NLHH198 pKa = 6.68LLFEE202 pKa = 5.28EE203 pKa = 4.66YY204 pKa = 10.86TNIPVSLLFNEE215 pKa = 5.05KK216 pKa = 9.61EE217 pKa = 4.34ANDD220 pKa = 4.02TDD222 pKa = 4.35HH223 pKa = 7.18EE224 pKa = 4.3PQISPGGLEE233 pKa = 4.2PPAEE237 pKa = 3.88KK238 pKa = 10.27HH239 pKa = 5.35FRR241 pKa = 11.84EE242 pKa = 4.52TEE244 pKa = 3.99VV245 pKa = 3.0
MM1 pKa = 7.74RR2 pKa = 11.84KK3 pKa = 9.61YY4 pKa = 10.63SPFRR8 pKa = 11.84NGYY11 pKa = 7.91MEE13 pKa = 4.35PTLGQHH19 pKa = 6.94LPTLSFPDD27 pKa = 3.77PGLRR31 pKa = 11.84PQNLYY36 pKa = 8.25TLWGDD41 pKa = 4.07SVVCLYY47 pKa = 10.65LYY49 pKa = 10.33QLSPPITWPLPPHH62 pKa = 7.02VIFCHH67 pKa = 6.21PGQLGAFLTNVPYY80 pKa = 10.94KK81 pKa = 10.66RR82 pKa = 11.84MEE84 pKa = 3.93EE85 pKa = 3.84LLYY88 pKa = 10.66KK89 pKa = 10.55ISLTTGALIILPEE102 pKa = 4.78DD103 pKa = 3.97CLPTTLFQPARR114 pKa = 11.84APVTLTAWQNGLLPFHH130 pKa = 6.75STLTTPGLIWTFTDD144 pKa = 3.88GTPMVSGPCPKK155 pKa = 10.26DD156 pKa = 3.49GQPSLVLQSSSFIFHH171 pKa = 7.35KK172 pKa = 10.59FQTKK176 pKa = 9.83AYY178 pKa = 8.81HH179 pKa = 6.37PSFLLSHH186 pKa = 6.6GLIQYY191 pKa = 10.76SSFHH195 pKa = 5.98NLHH198 pKa = 6.68LLFEE202 pKa = 5.28EE203 pKa = 4.66YY204 pKa = 10.86TNIPVSLLFNEE215 pKa = 5.05KK216 pKa = 9.61EE217 pKa = 4.34ANDD220 pKa = 4.02TDD222 pKa = 4.35HH223 pKa = 7.18EE224 pKa = 4.3PQISPGGLEE233 pKa = 4.2PPAEE237 pKa = 3.88KK238 pKa = 10.27HH239 pKa = 5.35FRR241 pKa = 11.84EE242 pKa = 4.52TEE244 pKa = 3.99VV245 pKa = 3.0
Molecular weight: 27.65 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q9WS54|Q9WS54_9STL1 Protease OS=Simian T-lymphotropic virus 1 OX=33747 PE=4 SV=1
MM1 pKa = 6.99QLAHH5 pKa = 7.39ILQPIRR11 pKa = 11.84QAFSQCTILQYY22 pKa = 10.34MDD24 pKa = 6.42DD25 pKa = 4.46ILLASPSHH33 pKa = 7.19KK34 pKa = 10.39DD35 pKa = 3.33LQLLSEE41 pKa = 4.32VTMASLISHH50 pKa = 7.03GLPVSEE56 pKa = 5.77NKK58 pKa = 8.15TQQTPGTIKK67 pKa = 10.65FLGQIISPNHH77 pKa = 4.96LTYY80 pKa = 10.85DD81 pKa = 3.91AVPTVPIRR89 pKa = 11.84SRR91 pKa = 11.84WALPEE96 pKa = 3.93LQALLGEE103 pKa = 4.47IQWVSKK109 pKa = 8.24GTPTLRR115 pKa = 11.84QPLHH119 pKa = 6.34SLYY122 pKa = 10.66CALQRR127 pKa = 11.84HH128 pKa = 5.66TDD130 pKa = 3.46PRR132 pKa = 11.84DD133 pKa = 3.62QICLNPSQVQSLVQLRR149 pKa = 11.84QALSQNCRR157 pKa = 11.84SRR159 pKa = 11.84LVQTVPLLGLVMLTLTGTTTVVFQSKK185 pKa = 7.33QQWPLVWLHH194 pKa = 6.78APLPHH199 pKa = 6.99TSQCPWGQLLASAVLLLDD217 pKa = 4.16KK218 pKa = 10.45YY219 pKa = 8.72TLQSYY224 pKa = 10.23GLLCQTIHH232 pKa = 6.81HH233 pKa = 6.74NISTQTFDD241 pKa = 4.64QFIQTSDD248 pKa = 3.57HH249 pKa = 6.49PSVPILYY256 pKa = 9.26HH257 pKa = 5.76HH258 pKa = 6.23SHH260 pKa = 6.6RR261 pKa = 11.84FKK263 pKa = 11.3NLGAQTGEE271 pKa = 3.76LWNTFLKK278 pKa = 9.77TAAPLAPVKK287 pKa = 10.77ALMPMFTLSPVIINAAPCLFSDD309 pKa = 4.86GSTSRR314 pKa = 11.84AAYY317 pKa = 9.42ILWDD321 pKa = 3.53KK322 pKa = 11.02HH323 pKa = 6.54ILSQKK328 pKa = 10.16SFPLPPPHH336 pKa = 7.31KK337 pKa = 9.2SAQRR341 pKa = 11.84AEE343 pKa = 4.27LLGLLHH349 pKa = 6.71GLSSAHH355 pKa = 6.23SWRR358 pKa = 11.84CLNIFLDD365 pKa = 4.03SKK367 pKa = 11.18YY368 pKa = 10.2LYY370 pKa = 10.25HH371 pKa = 6.84YY372 pKa = 10.73LRR374 pKa = 11.84TLALGTFQGRR384 pKa = 11.84SSQAPFQDD392 pKa = 3.7PLPRR396 pKa = 11.84LLSRR400 pKa = 11.84KK401 pKa = 8.61VVYY404 pKa = 9.83LHH406 pKa = 6.55HH407 pKa = 6.57VRR409 pKa = 11.84SHH411 pKa = 6.33TNLPDD416 pKa = 5.24PITRR420 pKa = 11.84LNALTDD426 pKa = 3.78ALLITPVLQLSPAEE440 pKa = 4.08LHH442 pKa = 6.79SLTHH446 pKa = 6.75CGQAALTLQGATTTEE461 pKa = 3.61ASNILRR467 pKa = 11.84SCHH470 pKa = 5.84ACRR473 pKa = 11.84KK474 pKa = 9.68NNPQHH479 pKa = 5.62QMPRR483 pKa = 11.84GHH485 pKa = 6.92IRR487 pKa = 11.84RR488 pKa = 11.84GLLPNHH494 pKa = 6.78IWQGDD499 pKa = 3.53ITHH502 pKa = 7.09FKK504 pKa = 10.36YY505 pKa = 10.9KK506 pKa = 8.54NTLYY510 pKa = 10.61RR511 pKa = 11.84LHH513 pKa = 6.88VWVDD517 pKa = 3.4TFSGAISATQKK528 pKa = 10.74KK529 pKa = 9.76KK530 pKa = 8.78EE531 pKa = 4.25TSSEE535 pKa = 4.27AISSLLQAIAYY546 pKa = 8.61LGKK549 pKa = 9.73PNYY552 pKa = 9.76INTDD556 pKa = 3.0NGPAYY561 pKa = 9.83ISQDD565 pKa = 3.65FLDD568 pKa = 4.16MCTPLAIRR576 pKa = 11.84HH577 pKa = 4.98TTHH580 pKa = 5.93VPYY583 pKa = 10.85NPTSSGLVEE592 pKa = 4.4RR593 pKa = 11.84SNGILKK599 pKa = 8.91TLLYY603 pKa = 10.45KK604 pKa = 10.93YY605 pKa = 9.32FTDD608 pKa = 5.39KK609 pKa = 10.95PDD611 pKa = 3.67LPMDD615 pKa = 4.1NALSIALWTINHH627 pKa = 6.38LNVLTHH633 pKa = 5.36CHH635 pKa = 4.4KK636 pKa = 9.75TRR638 pKa = 11.84WQLHH642 pKa = 5.27HH643 pKa = 6.85SPRR646 pKa = 11.84LQPIPEE652 pKa = 4.12TRR654 pKa = 11.84SLSNKK659 pKa = 5.47QTHH662 pKa = 5.75WYY664 pKa = 8.91YY665 pKa = 11.25FKK667 pKa = 11.28LPGLNSRR674 pKa = 11.84QWKK677 pKa = 9.58GPQEE681 pKa = 4.28ALQEE685 pKa = 4.25AAGPALIPVIASSAQWIPWRR705 pKa = 11.84LLKK708 pKa = 10.48RR709 pKa = 11.84AACPRR714 pKa = 11.84PVGAPPIPKK723 pKa = 10.07KK724 pKa = 9.95KK725 pKa = 7.01TTNTMGKK732 pKa = 9.76FLTTLILFFQFCPLILGDD750 pKa = 4.58YY751 pKa = 10.57SPSCCTLTIGVSSYY765 pKa = 10.86HH766 pKa = 6.73SKK768 pKa = 10.17PCNPAQPVCSWTLDD782 pKa = 3.88LLALSADD789 pKa = 3.67QALQPPCPNLVSYY802 pKa = 10.82SSYY805 pKa = 9.9HH806 pKa = 5.03ATYY809 pKa = 10.81SLYY812 pKa = 10.83LFPHH816 pKa = 7.5WIKK819 pKa = 10.78KK820 pKa = 8.39PNRR823 pKa = 11.84NGGGYY828 pKa = 10.33YY829 pKa = 9.83SASYY833 pKa = 10.97SDD835 pKa = 4.19PCSLKK840 pKa = 10.57CPYY843 pKa = 10.0LGCQSWTCPYY853 pKa = 9.29TGAISSPYY861 pKa = 9.66WKK863 pKa = 10.1FQQDD867 pKa = 3.56VNFTQEE873 pKa = 3.84VSRR876 pKa = 11.84LTINLHH882 pKa = 5.47FSKK885 pKa = 10.77CGFPFSLLIDD895 pKa = 3.93APGYY899 pKa = 10.57DD900 pKa = 4.9PIWFLNTEE908 pKa = 4.47PSQLPPTAPPLLPHH922 pKa = 6.84SNLDD926 pKa = 3.99HH927 pKa = 7.3ILEE930 pKa = 4.41PSIPWKK936 pKa = 10.74SKK938 pKa = 10.61LLTPVQLTLQSTNYY952 pKa = 8.58TCIVCIDD959 pKa = 3.86RR960 pKa = 11.84ASLSTWHH967 pKa = 6.34VLYY970 pKa = 10.3FPNVSVPSSPSTPLLYY986 pKa = 10.08PSLALPAPHH995 pKa = 6.68LTLPFNWTHH1004 pKa = 6.49CFNPQIQAIVSSPCHH1019 pKa = 6.19NSLILPPFSLSPVPTLGSRR1038 pKa = 11.84SRR1040 pKa = 11.84RR1041 pKa = 11.84AVPVAVWLVSALAMGAGMAGGITGSMSLASGRR1073 pKa = 11.84SLLHH1077 pKa = 6.58EE1078 pKa = 4.11VDD1080 pKa = 4.21KK1081 pKa = 11.3DD1082 pKa = 3.49ISQLTQAIVKK1092 pKa = 8.27NHH1094 pKa = 6.09KK1095 pKa = 9.92NLLKK1099 pKa = 9.97IAQYY1103 pKa = 10.57AAQNRR1108 pKa = 11.84RR1109 pKa = 11.84GLDD1112 pKa = 3.36LLFWEE1117 pKa = 5.19QGGLCKK1123 pKa = 10.25ALQEE1127 pKa = 4.27QCCFLNITNSHH1138 pKa = 4.81VSILQEE1144 pKa = 4.04RR1145 pKa = 11.84PPLEE1149 pKa = 4.01NRR1151 pKa = 11.84VLTGWGLNWDD1161 pKa = 4.35LGLSQWARR1169 pKa = 11.84EE1170 pKa = 3.91ALQTGITLVALLPLVILAGPCILRR1194 pKa = 11.84QLRR1197 pKa = 11.84HH1198 pKa = 5.3LPSRR1202 pKa = 11.84VRR1204 pKa = 11.84HH1205 pKa = 4.99PHH1207 pKa = 5.44YY1208 pKa = 11.08SLINPEE1214 pKa = 4.01SCLL1217 pKa = 3.69
MM1 pKa = 6.99QLAHH5 pKa = 7.39ILQPIRR11 pKa = 11.84QAFSQCTILQYY22 pKa = 10.34MDD24 pKa = 6.42DD25 pKa = 4.46ILLASPSHH33 pKa = 7.19KK34 pKa = 10.39DD35 pKa = 3.33LQLLSEE41 pKa = 4.32VTMASLISHH50 pKa = 7.03GLPVSEE56 pKa = 5.77NKK58 pKa = 8.15TQQTPGTIKK67 pKa = 10.65FLGQIISPNHH77 pKa = 4.96LTYY80 pKa = 10.85DD81 pKa = 3.91AVPTVPIRR89 pKa = 11.84SRR91 pKa = 11.84WALPEE96 pKa = 3.93LQALLGEE103 pKa = 4.47IQWVSKK109 pKa = 8.24GTPTLRR115 pKa = 11.84QPLHH119 pKa = 6.34SLYY122 pKa = 10.66CALQRR127 pKa = 11.84HH128 pKa = 5.66TDD130 pKa = 3.46PRR132 pKa = 11.84DD133 pKa = 3.62QICLNPSQVQSLVQLRR149 pKa = 11.84QALSQNCRR157 pKa = 11.84SRR159 pKa = 11.84LVQTVPLLGLVMLTLTGTTTVVFQSKK185 pKa = 7.33QQWPLVWLHH194 pKa = 6.78APLPHH199 pKa = 6.99TSQCPWGQLLASAVLLLDD217 pKa = 4.16KK218 pKa = 10.45YY219 pKa = 8.72TLQSYY224 pKa = 10.23GLLCQTIHH232 pKa = 6.81HH233 pKa = 6.74NISTQTFDD241 pKa = 4.64QFIQTSDD248 pKa = 3.57HH249 pKa = 6.49PSVPILYY256 pKa = 9.26HH257 pKa = 5.76HH258 pKa = 6.23SHH260 pKa = 6.6RR261 pKa = 11.84FKK263 pKa = 11.3NLGAQTGEE271 pKa = 3.76LWNTFLKK278 pKa = 9.77TAAPLAPVKK287 pKa = 10.77ALMPMFTLSPVIINAAPCLFSDD309 pKa = 4.86GSTSRR314 pKa = 11.84AAYY317 pKa = 9.42ILWDD321 pKa = 3.53KK322 pKa = 11.02HH323 pKa = 6.54ILSQKK328 pKa = 10.16SFPLPPPHH336 pKa = 7.31KK337 pKa = 9.2SAQRR341 pKa = 11.84AEE343 pKa = 4.27LLGLLHH349 pKa = 6.71GLSSAHH355 pKa = 6.23SWRR358 pKa = 11.84CLNIFLDD365 pKa = 4.03SKK367 pKa = 11.18YY368 pKa = 10.2LYY370 pKa = 10.25HH371 pKa = 6.84YY372 pKa = 10.73LRR374 pKa = 11.84TLALGTFQGRR384 pKa = 11.84SSQAPFQDD392 pKa = 3.7PLPRR396 pKa = 11.84LLSRR400 pKa = 11.84KK401 pKa = 8.61VVYY404 pKa = 9.83LHH406 pKa = 6.55HH407 pKa = 6.57VRR409 pKa = 11.84SHH411 pKa = 6.33TNLPDD416 pKa = 5.24PITRR420 pKa = 11.84LNALTDD426 pKa = 3.78ALLITPVLQLSPAEE440 pKa = 4.08LHH442 pKa = 6.79SLTHH446 pKa = 6.75CGQAALTLQGATTTEE461 pKa = 3.61ASNILRR467 pKa = 11.84SCHH470 pKa = 5.84ACRR473 pKa = 11.84KK474 pKa = 9.68NNPQHH479 pKa = 5.62QMPRR483 pKa = 11.84GHH485 pKa = 6.92IRR487 pKa = 11.84RR488 pKa = 11.84GLLPNHH494 pKa = 6.78IWQGDD499 pKa = 3.53ITHH502 pKa = 7.09FKK504 pKa = 10.36YY505 pKa = 10.9KK506 pKa = 8.54NTLYY510 pKa = 10.61RR511 pKa = 11.84LHH513 pKa = 6.88VWVDD517 pKa = 3.4TFSGAISATQKK528 pKa = 10.74KK529 pKa = 9.76KK530 pKa = 8.78EE531 pKa = 4.25TSSEE535 pKa = 4.27AISSLLQAIAYY546 pKa = 8.61LGKK549 pKa = 9.73PNYY552 pKa = 9.76INTDD556 pKa = 3.0NGPAYY561 pKa = 9.83ISQDD565 pKa = 3.65FLDD568 pKa = 4.16MCTPLAIRR576 pKa = 11.84HH577 pKa = 4.98TTHH580 pKa = 5.93VPYY583 pKa = 10.85NPTSSGLVEE592 pKa = 4.4RR593 pKa = 11.84SNGILKK599 pKa = 8.91TLLYY603 pKa = 10.45KK604 pKa = 10.93YY605 pKa = 9.32FTDD608 pKa = 5.39KK609 pKa = 10.95PDD611 pKa = 3.67LPMDD615 pKa = 4.1NALSIALWTINHH627 pKa = 6.38LNVLTHH633 pKa = 5.36CHH635 pKa = 4.4KK636 pKa = 9.75TRR638 pKa = 11.84WQLHH642 pKa = 5.27HH643 pKa = 6.85SPRR646 pKa = 11.84LQPIPEE652 pKa = 4.12TRR654 pKa = 11.84SLSNKK659 pKa = 5.47QTHH662 pKa = 5.75WYY664 pKa = 8.91YY665 pKa = 11.25FKK667 pKa = 11.28LPGLNSRR674 pKa = 11.84QWKK677 pKa = 9.58GPQEE681 pKa = 4.28ALQEE685 pKa = 4.25AAGPALIPVIASSAQWIPWRR705 pKa = 11.84LLKK708 pKa = 10.48RR709 pKa = 11.84AACPRR714 pKa = 11.84PVGAPPIPKK723 pKa = 10.07KK724 pKa = 9.95KK725 pKa = 7.01TTNTMGKK732 pKa = 9.76FLTTLILFFQFCPLILGDD750 pKa = 4.58YY751 pKa = 10.57SPSCCTLTIGVSSYY765 pKa = 10.86HH766 pKa = 6.73SKK768 pKa = 10.17PCNPAQPVCSWTLDD782 pKa = 3.88LLALSADD789 pKa = 3.67QALQPPCPNLVSYY802 pKa = 10.82SSYY805 pKa = 9.9HH806 pKa = 5.03ATYY809 pKa = 10.81SLYY812 pKa = 10.83LFPHH816 pKa = 7.5WIKK819 pKa = 10.78KK820 pKa = 8.39PNRR823 pKa = 11.84NGGGYY828 pKa = 10.33YY829 pKa = 9.83SASYY833 pKa = 10.97SDD835 pKa = 4.19PCSLKK840 pKa = 10.57CPYY843 pKa = 10.0LGCQSWTCPYY853 pKa = 9.29TGAISSPYY861 pKa = 9.66WKK863 pKa = 10.1FQQDD867 pKa = 3.56VNFTQEE873 pKa = 3.84VSRR876 pKa = 11.84LTINLHH882 pKa = 5.47FSKK885 pKa = 10.77CGFPFSLLIDD895 pKa = 3.93APGYY899 pKa = 10.57DD900 pKa = 4.9PIWFLNTEE908 pKa = 4.47PSQLPPTAPPLLPHH922 pKa = 6.84SNLDD926 pKa = 3.99HH927 pKa = 7.3ILEE930 pKa = 4.41PSIPWKK936 pKa = 10.74SKK938 pKa = 10.61LLTPVQLTLQSTNYY952 pKa = 8.58TCIVCIDD959 pKa = 3.86RR960 pKa = 11.84ASLSTWHH967 pKa = 6.34VLYY970 pKa = 10.3FPNVSVPSSPSTPLLYY986 pKa = 10.08PSLALPAPHH995 pKa = 6.68LTLPFNWTHH1004 pKa = 6.49CFNPQIQAIVSSPCHH1019 pKa = 6.19NSLILPPFSLSPVPTLGSRR1038 pKa = 11.84SRR1040 pKa = 11.84RR1041 pKa = 11.84AVPVAVWLVSALAMGAGMAGGITGSMSLASGRR1073 pKa = 11.84SLLHH1077 pKa = 6.58EE1078 pKa = 4.11VDD1080 pKa = 4.21KK1081 pKa = 11.3DD1082 pKa = 3.49ISQLTQAIVKK1092 pKa = 8.27NHH1094 pKa = 6.09KK1095 pKa = 9.92NLLKK1099 pKa = 9.97IAQYY1103 pKa = 10.57AAQNRR1108 pKa = 11.84RR1109 pKa = 11.84GLDD1112 pKa = 3.36LLFWEE1117 pKa = 5.19QGGLCKK1123 pKa = 10.25ALQEE1127 pKa = 4.27QCCFLNITNSHH1138 pKa = 4.81VSILQEE1144 pKa = 4.04RR1145 pKa = 11.84PPLEE1149 pKa = 4.01NRR1151 pKa = 11.84VLTGWGLNWDD1161 pKa = 4.35LGLSQWARR1169 pKa = 11.84EE1170 pKa = 3.91ALQTGITLVALLPLVILAGPCILRR1194 pKa = 11.84QLRR1197 pKa = 11.84HH1198 pKa = 5.3LPSRR1202 pKa = 11.84VRR1204 pKa = 11.84HH1205 pKa = 4.99PHH1207 pKa = 5.44YY1208 pKa = 11.08SLINPEE1214 pKa = 4.01SCLL1217 pKa = 3.69
Molecular weight: 135.83 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2056 |
166 |
1217 |
514.0 |
57.24 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.518 ± 0.966 | 2.529 ± 0.238 |
3.356 ± 0.557 | 2.918 ± 0.818 |
3.064 ± 0.614 | 5.01 ± 0.51 |
4.037 ± 0.619 | 5.01 ± 0.391 |
3.842 ± 0.329 | 13.765 ± 1.03 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.216 ± 0.13 | 3.696 ± 0.278 |
10.554 ± 1.727 | 6.469 ± 0.603 |
4.086 ± 0.381 | 8.123 ± 0.9 |
6.663 ± 1.109 | 4.037 ± 0.363 |
2.14 ± 0.212 | 2.967 ± 0.45 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
