
Tangfeifania diversioriginum
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Bacteroidia; Marinilabiliales; Prolixibacteraceae; Tangfeifania
Average proteome isoelectric point is 6.46
Get precalculated fractions of proteins
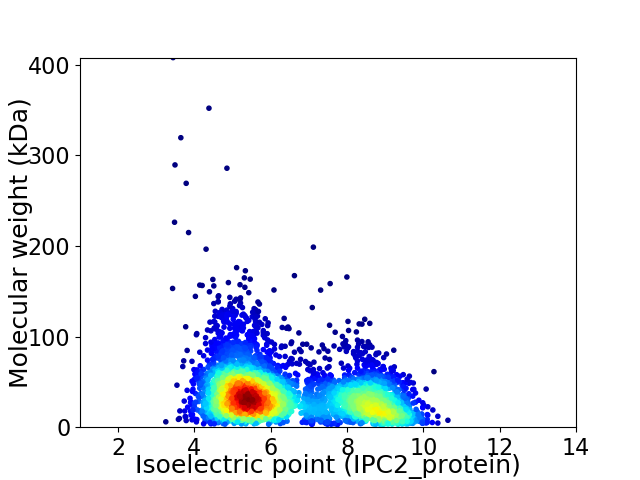
Virtual 2D-PAGE plot for 4103 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1M6H0C1|A0A1M6H0C1_9BACT Cupin domain-containing protein OS=Tangfeifania diversioriginum OX=1168035 GN=SAMN05444280_11234 PE=4 SV=1
MM1 pKa = 7.61KK2 pKa = 10.45KK3 pKa = 10.28LLLSFSVVLIFLFTGMQTTWAQATINVPTDD33 pKa = 3.05QPTIQDD39 pKa = 4.6AITAASANDD48 pKa = 4.2IINVAAGTYY57 pKa = 9.61NEE59 pKa = 4.83SFILDD64 pKa = 3.65KK65 pKa = 11.16TGLTLQGPNEE75 pKa = 4.05ALNGNDD81 pKa = 3.37AGRR84 pKa = 11.84AAEE87 pKa = 4.67AIIEE91 pKa = 4.24GVARR95 pKa = 11.84VEE97 pKa = 4.05ADD99 pKa = 3.35DD100 pKa = 5.04VIIKK104 pKa = 10.6GLVIDD109 pKa = 4.95GANVAQTTNLTMRR122 pKa = 11.84GILVANTNARR132 pKa = 11.84DD133 pKa = 3.53NVTIEE138 pKa = 3.81NNIIKK143 pKa = 10.21NWVTGISLAGSSTFAWVDD161 pKa = 3.16GATITGNLFVNNGVGSTEE179 pKa = 4.08NVDD182 pKa = 3.74GLSITNNAFDD192 pKa = 3.87NGGIGLGGGAVLAVPITGNDD212 pKa = 3.34FSNGSRR218 pKa = 11.84YY219 pKa = 9.45ISAATGVTADD229 pKa = 4.15FDD231 pKa = 4.68AMLSANTFDD240 pKa = 3.39GGAYY244 pKa = 10.27AEE246 pKa = 5.36LITGQWYY253 pKa = 9.63DD254 pKa = 3.23RR255 pKa = 11.84AIFSTIQGAIDD266 pKa = 3.32KK267 pKa = 10.62AGAGSVAKK275 pKa = 10.65VADD278 pKa = 3.49GTYY281 pKa = 10.4NEE283 pKa = 4.9NVTISDD289 pKa = 4.24NISLISEE296 pKa = 4.28NGASSTIIDD305 pKa = 3.91GDD307 pKa = 3.69NSGSEE312 pKa = 4.36LGTILLKK319 pKa = 10.79PGGNGISIGAVGQGFTIKK337 pKa = 10.88GIDD340 pKa = 3.62GPAGLEE346 pKa = 3.59KK347 pKa = 10.55AAIYY351 pKa = 9.14FQGAQDD357 pKa = 4.22NITIEE362 pKa = 4.16GNIIEE367 pKa = 4.53ARR369 pKa = 11.84GDD371 pKa = 3.36AALLGEE377 pKa = 4.43YY378 pKa = 9.99NAANNNITITNNEE391 pKa = 3.58ITGQTFTGANPAGVGFGAQFSLPNVPRR418 pKa = 11.84QLVTFGGGSGTTNTQNFTFTNNIISGIAGGMSITDD453 pKa = 3.76DD454 pKa = 3.65NGNTISPTEE463 pKa = 3.94QGNTMVTLDD472 pKa = 3.84LVGTNVITGNTFSGTTTRR490 pKa = 11.84YY491 pKa = 10.05AYY493 pKa = 10.46ALRR496 pKa = 11.84VRR498 pKa = 11.84GAGAYY503 pKa = 7.73TITEE507 pKa = 4.09NNFTGSYY514 pKa = 9.37PGFIFSDD521 pKa = 4.0SNPITATCNWFGTTDD536 pKa = 3.35EE537 pKa = 4.71SVISSKK543 pKa = 10.14MDD545 pKa = 3.33ASITFIPYY553 pKa = 10.26NISQGGACVGGLNYY567 pKa = 10.44AVNIEE572 pKa = 4.32SPLASSNCGTLDD584 pKa = 3.72VPVTVQDD591 pKa = 4.01FNNVGAISLRR601 pKa = 11.84LNYY604 pKa = 10.56DD605 pKa = 3.62DD606 pKa = 6.12VLLSYY611 pKa = 11.63NDD613 pKa = 3.34VTLNTAIKK621 pKa = 10.38SATQVGEE628 pKa = 3.96TGTQEE633 pKa = 4.32GEE635 pKa = 3.97NKK637 pKa = 10.16GQIRR641 pKa = 11.84LAYY644 pKa = 10.2FGDD647 pKa = 4.03AVTLNDD653 pKa = 4.57DD654 pKa = 3.97DD655 pKa = 5.34VLFTLHH661 pKa = 7.12FDD663 pKa = 3.67ILSSTAGNSTTDD675 pKa = 3.65LTWDD679 pKa = 3.43KK680 pKa = 11.13SSPGYY685 pKa = 10.42CEE687 pKa = 3.74LAGPGGTPVYY697 pKa = 10.05TSTFNDD703 pKa = 3.14VTGVVIPEE711 pKa = 3.94RR712 pKa = 11.84PVKK715 pKa = 10.72NVDD718 pKa = 2.9TGRR721 pKa = 11.84EE722 pKa = 3.82YY723 pKa = 11.28CKK725 pKa = 10.55IQDD728 pKa = 4.95AIDD731 pKa = 3.87ADD733 pKa = 4.04EE734 pKa = 4.55TDD736 pKa = 3.53NGDD739 pKa = 3.85VIEE742 pKa = 4.5VSAGEE747 pKa = 4.13YY748 pKa = 10.13VEE750 pKa = 5.44SVDD753 pKa = 3.71VYY755 pKa = 11.31KK756 pKa = 10.98SVILKK761 pKa = 10.52GADD764 pKa = 3.3KK765 pKa = 11.2NSTFIQAPSTLPAASDD781 pKa = 3.46ALSNIVYY788 pKa = 9.36VHH790 pKa = 6.62GSGISTEE797 pKa = 3.74ISGFTIEE804 pKa = 4.85GPGPSGCGSIGRR816 pKa = 11.84GIFVSDD822 pKa = 3.55GAYY825 pKa = 10.76ANIHH829 pKa = 6.98DD830 pKa = 4.77NNILDD835 pKa = 3.59IRR837 pKa = 11.84DD838 pKa = 3.79NPFSGCQNGIAIQIGRR854 pKa = 11.84NALSSSGTAIIHH866 pKa = 5.78NNIISGYY873 pKa = 7.77QKK875 pKa = 10.87GAIVVDD881 pKa = 3.8NSGSNATITNNTITGTGTTDD901 pKa = 2.72VTAQNGIQVSRR912 pKa = 11.84GATATLGGNTISGNSFHH929 pKa = 7.46EE930 pKa = 5.31EE931 pKa = 3.81GSQWDD936 pKa = 3.41WGACGILLYY945 pKa = 10.71QSGAVSLTGGNNISGNDD962 pKa = 3.51QNYY965 pKa = 8.75YY966 pKa = 10.84AYY968 pKa = 10.34DD969 pKa = 3.41VAGNLILGAEE979 pKa = 4.3TFGASTAPVTFGYY992 pKa = 10.78NIADD996 pKa = 3.69YY997 pKa = 10.44TNQNIDD1003 pKa = 3.07ASQCTFAEE1011 pKa = 4.66GNPASAVLSEE1021 pKa = 4.74LFDD1024 pKa = 4.48IEE1026 pKa = 4.36DD1027 pKa = 4.87RR1028 pKa = 11.84IWHH1031 pKa = 5.99SVDD1034 pKa = 3.3DD1035 pKa = 3.92QTISGFVNIKK1045 pKa = 10.04AGNVYY1050 pKa = 8.47VTHH1053 pKa = 6.77TEE1055 pKa = 3.91SGAKK1059 pKa = 8.36IQYY1062 pKa = 10.2GIDD1065 pKa = 3.32AASVNDD1071 pKa = 3.87VIHH1074 pKa = 6.25VKK1076 pKa = 10.5EE1077 pKa = 4.19GTYY1080 pKa = 10.35SEE1082 pKa = 5.19NITVDD1087 pKa = 3.08KK1088 pKa = 11.09DD1089 pKa = 3.64VTLLGANAGVACDD1102 pKa = 3.64TRR1104 pKa = 11.84GAEE1107 pKa = 4.09STIAGTGGAAVSVASDD1123 pKa = 3.41GVTIDD1128 pKa = 3.61GFEE1131 pKa = 4.01ITNPTGNYY1139 pKa = 9.39AVNSTGNNNLLLQYY1153 pKa = 11.37NNVNDD1158 pKa = 3.82VGTSGTPGNVHH1169 pKa = 6.66AVAISMGSANTDD1181 pKa = 3.16DD1182 pKa = 5.51VKK1184 pKa = 10.82IQHH1187 pKa = 6.1NCISNINNAGNTHH1200 pKa = 6.35SGSGIAVGFSTATTDD1215 pKa = 3.11ITNLYY1220 pKa = 9.24IGYY1223 pKa = 8.29NTISQVNSNNALDD1236 pKa = 3.91YY1237 pKa = 11.25ASGGRR1242 pKa = 11.84GANGIILNVGANSTGKK1258 pKa = 8.41VTDD1261 pKa = 3.97GVVEE1265 pKa = 4.23YY1266 pKa = 9.08NTIHH1270 pKa = 7.15DD1271 pKa = 4.02LSGYY1275 pKa = 7.27WVHH1278 pKa = 6.89GVGLEE1283 pKa = 4.14GNTPGALVQNNLIYY1297 pKa = 10.88NLVSTYY1303 pKa = 10.37TDD1305 pKa = 2.93IPGVGVKK1312 pKa = 10.3VEE1314 pKa = 4.74DD1315 pKa = 3.5NTGAATVSVNNNSFTSLSPGILNAVSGATVNATCNWFAADD1355 pKa = 3.69VTEE1358 pKa = 4.87KK1359 pKa = 11.01SLGDD1363 pKa = 3.5VTVVPWLVDD1372 pKa = 3.31GTNSVASGPGFEE1384 pKa = 4.71PATACVACAISIDD1397 pKa = 3.64AVVTNVSCPGGADD1410 pKa = 3.49DD1411 pKa = 5.16GAIDD1415 pKa = 3.63VTVTNGVSPYY1425 pKa = 8.55TYY1427 pKa = 10.48SWTGPDD1433 pKa = 4.07GFTSPDD1439 pKa = 3.24EE1440 pKa = 5.24DD1441 pKa = 3.76ISGLAAGTYY1450 pKa = 9.97NLLVTADD1457 pKa = 3.82NGCSKK1462 pKa = 10.72EE1463 pKa = 4.0KK1464 pKa = 10.65VVVVGTDD1471 pKa = 3.19ADD1473 pKa = 3.95VTAPTITEE1481 pKa = 4.02ALDD1484 pKa = 3.21VLTEE1488 pKa = 4.13EE1489 pKa = 5.15GCTDD1493 pKa = 3.69ADD1495 pKa = 3.81VPVAKK1500 pKa = 8.37TTVAEE1505 pKa = 4.16LEE1507 pKa = 4.2AMGVIIDD1514 pKa = 5.1DD1515 pKa = 4.0CTLDD1519 pKa = 3.44EE1520 pKa = 4.8NMTVSYY1526 pKa = 10.93SDD1528 pKa = 4.14ASAGTCPVVVTRR1540 pKa = 11.84TYY1542 pKa = 10.86TVTDD1546 pKa = 3.91ASNNSVTVDD1555 pKa = 3.1QTINIEE1561 pKa = 4.38DD1562 pKa = 3.77NTPPSIACPPPIAVKK1577 pKa = 10.4MNDD1580 pKa = 2.96GCEE1583 pKa = 3.83YY1584 pKa = 11.03DD1585 pKa = 3.63PAIGSGIGVATATDD1599 pKa = 3.62NCSDD1603 pKa = 3.87AVDD1606 pKa = 3.55IQISNDD1612 pKa = 2.49ITGNLTEE1619 pKa = 4.8GEE1621 pKa = 4.26NFITWTAVDD1630 pKa = 3.66EE1631 pKa = 4.74CNNSVTCTQKK1641 pKa = 9.23VTVIRR1646 pKa = 11.84NTLSGTVMYY1655 pKa = 10.62NNTAQTPMANVVLKK1669 pKa = 11.06LFDD1672 pKa = 4.61SNDD1675 pKa = 3.6DD1676 pKa = 3.59QVGDD1680 pKa = 3.98DD1681 pKa = 3.8VVTSEE1686 pKa = 4.35TNPGNFEE1693 pKa = 4.06FTGLCAGDD1701 pKa = 3.8YY1702 pKa = 9.97TVVASDD1708 pKa = 3.56TSKK1711 pKa = 10.92VGYY1714 pKa = 10.31INATDD1719 pKa = 3.64AGAANAWGTAGGDD1732 pKa = 3.11IEE1734 pKa = 4.88YY1735 pKa = 10.69VQFLAGDD1742 pKa = 3.66VNDD1745 pKa = 4.07NLLINSTDD1753 pKa = 3.09AQAIQNYY1760 pKa = 8.26FVFGGSFTKK1769 pKa = 10.04PWEE1772 pKa = 3.9YY1773 pKa = 10.25WSAGSVIHH1781 pKa = 6.85SNYY1784 pKa = 10.33NPFSSNAIPPHH1795 pKa = 5.91PAAIEE1800 pKa = 4.13VVISGGDD1807 pKa = 3.51VTHH1810 pKa = 7.5DD1811 pKa = 4.84LYY1813 pKa = 11.59AQATGDD1819 pKa = 3.8FNGSFGPTLKK1829 pKa = 10.73SAGSSVRR1836 pKa = 11.84LSADD1840 pKa = 2.77RR1841 pKa = 11.84NMNVEE1846 pKa = 4.15EE1847 pKa = 4.06NQVFNLPLRR1856 pKa = 11.84SEE1858 pKa = 4.37FAMEE1862 pKa = 4.04VGAVSMILDD1871 pKa = 3.62MPEE1874 pKa = 4.81DD1875 pKa = 4.05VVQVTGVQIAGSEE1888 pKa = 4.49VPVTWAMKK1896 pKa = 10.45DD1897 pKa = 3.13NEE1899 pKa = 5.07LRR1901 pKa = 11.84IGWNSLNPVYY1911 pKa = 10.75VGEE1914 pKa = 4.17NDD1916 pKa = 3.26AFVVLKK1922 pKa = 10.47LQTSANFTEE1931 pKa = 4.62GQLMDD1936 pKa = 3.85IGLVYY1941 pKa = 11.02NPLNEE1946 pKa = 4.13LADD1949 pKa = 4.07GNFEE1953 pKa = 4.76PIEE1956 pKa = 4.1NAALKK1961 pKa = 9.53VARR1964 pKa = 11.84VGNGLTDD1971 pKa = 3.22INEE1974 pKa = 4.59DD1975 pKa = 3.46LDD1977 pKa = 3.75IDD1979 pKa = 4.35KK1980 pKa = 8.13MTFSNYY1986 pKa = 9.15PNPFSEE1992 pKa = 4.56STTLMYY1998 pKa = 8.58TIPVDD2003 pKa = 3.5GKK2005 pKa = 11.33VNISVYY2011 pKa = 10.38NALGQLVTTIVDD2023 pKa = 3.35EE2024 pKa = 4.23NQRR2027 pKa = 11.84TGRR2030 pKa = 11.84YY2031 pKa = 7.14TINNCGDD2038 pKa = 3.47EE2039 pKa = 4.74LQPGMYY2045 pKa = 9.15ISKK2048 pKa = 10.67LMLTNGTEE2056 pKa = 4.41SMVSTIKK2063 pKa = 11.02LNVVKK2068 pKa = 10.82
MM1 pKa = 7.61KK2 pKa = 10.45KK3 pKa = 10.28LLLSFSVVLIFLFTGMQTTWAQATINVPTDD33 pKa = 3.05QPTIQDD39 pKa = 4.6AITAASANDD48 pKa = 4.2IINVAAGTYY57 pKa = 9.61NEE59 pKa = 4.83SFILDD64 pKa = 3.65KK65 pKa = 11.16TGLTLQGPNEE75 pKa = 4.05ALNGNDD81 pKa = 3.37AGRR84 pKa = 11.84AAEE87 pKa = 4.67AIIEE91 pKa = 4.24GVARR95 pKa = 11.84VEE97 pKa = 4.05ADD99 pKa = 3.35DD100 pKa = 5.04VIIKK104 pKa = 10.6GLVIDD109 pKa = 4.95GANVAQTTNLTMRR122 pKa = 11.84GILVANTNARR132 pKa = 11.84DD133 pKa = 3.53NVTIEE138 pKa = 3.81NNIIKK143 pKa = 10.21NWVTGISLAGSSTFAWVDD161 pKa = 3.16GATITGNLFVNNGVGSTEE179 pKa = 4.08NVDD182 pKa = 3.74GLSITNNAFDD192 pKa = 3.87NGGIGLGGGAVLAVPITGNDD212 pKa = 3.34FSNGSRR218 pKa = 11.84YY219 pKa = 9.45ISAATGVTADD229 pKa = 4.15FDD231 pKa = 4.68AMLSANTFDD240 pKa = 3.39GGAYY244 pKa = 10.27AEE246 pKa = 5.36LITGQWYY253 pKa = 9.63DD254 pKa = 3.23RR255 pKa = 11.84AIFSTIQGAIDD266 pKa = 3.32KK267 pKa = 10.62AGAGSVAKK275 pKa = 10.65VADD278 pKa = 3.49GTYY281 pKa = 10.4NEE283 pKa = 4.9NVTISDD289 pKa = 4.24NISLISEE296 pKa = 4.28NGASSTIIDD305 pKa = 3.91GDD307 pKa = 3.69NSGSEE312 pKa = 4.36LGTILLKK319 pKa = 10.79PGGNGISIGAVGQGFTIKK337 pKa = 10.88GIDD340 pKa = 3.62GPAGLEE346 pKa = 3.59KK347 pKa = 10.55AAIYY351 pKa = 9.14FQGAQDD357 pKa = 4.22NITIEE362 pKa = 4.16GNIIEE367 pKa = 4.53ARR369 pKa = 11.84GDD371 pKa = 3.36AALLGEE377 pKa = 4.43YY378 pKa = 9.99NAANNNITITNNEE391 pKa = 3.58ITGQTFTGANPAGVGFGAQFSLPNVPRR418 pKa = 11.84QLVTFGGGSGTTNTQNFTFTNNIISGIAGGMSITDD453 pKa = 3.76DD454 pKa = 3.65NGNTISPTEE463 pKa = 3.94QGNTMVTLDD472 pKa = 3.84LVGTNVITGNTFSGTTTRR490 pKa = 11.84YY491 pKa = 10.05AYY493 pKa = 10.46ALRR496 pKa = 11.84VRR498 pKa = 11.84GAGAYY503 pKa = 7.73TITEE507 pKa = 4.09NNFTGSYY514 pKa = 9.37PGFIFSDD521 pKa = 4.0SNPITATCNWFGTTDD536 pKa = 3.35EE537 pKa = 4.71SVISSKK543 pKa = 10.14MDD545 pKa = 3.33ASITFIPYY553 pKa = 10.26NISQGGACVGGLNYY567 pKa = 10.44AVNIEE572 pKa = 4.32SPLASSNCGTLDD584 pKa = 3.72VPVTVQDD591 pKa = 4.01FNNVGAISLRR601 pKa = 11.84LNYY604 pKa = 10.56DD605 pKa = 3.62DD606 pKa = 6.12VLLSYY611 pKa = 11.63NDD613 pKa = 3.34VTLNTAIKK621 pKa = 10.38SATQVGEE628 pKa = 3.96TGTQEE633 pKa = 4.32GEE635 pKa = 3.97NKK637 pKa = 10.16GQIRR641 pKa = 11.84LAYY644 pKa = 10.2FGDD647 pKa = 4.03AVTLNDD653 pKa = 4.57DD654 pKa = 3.97DD655 pKa = 5.34VLFTLHH661 pKa = 7.12FDD663 pKa = 3.67ILSSTAGNSTTDD675 pKa = 3.65LTWDD679 pKa = 3.43KK680 pKa = 11.13SSPGYY685 pKa = 10.42CEE687 pKa = 3.74LAGPGGTPVYY697 pKa = 10.05TSTFNDD703 pKa = 3.14VTGVVIPEE711 pKa = 3.94RR712 pKa = 11.84PVKK715 pKa = 10.72NVDD718 pKa = 2.9TGRR721 pKa = 11.84EE722 pKa = 3.82YY723 pKa = 11.28CKK725 pKa = 10.55IQDD728 pKa = 4.95AIDD731 pKa = 3.87ADD733 pKa = 4.04EE734 pKa = 4.55TDD736 pKa = 3.53NGDD739 pKa = 3.85VIEE742 pKa = 4.5VSAGEE747 pKa = 4.13YY748 pKa = 10.13VEE750 pKa = 5.44SVDD753 pKa = 3.71VYY755 pKa = 11.31KK756 pKa = 10.98SVILKK761 pKa = 10.52GADD764 pKa = 3.3KK765 pKa = 11.2NSTFIQAPSTLPAASDD781 pKa = 3.46ALSNIVYY788 pKa = 9.36VHH790 pKa = 6.62GSGISTEE797 pKa = 3.74ISGFTIEE804 pKa = 4.85GPGPSGCGSIGRR816 pKa = 11.84GIFVSDD822 pKa = 3.55GAYY825 pKa = 10.76ANIHH829 pKa = 6.98DD830 pKa = 4.77NNILDD835 pKa = 3.59IRR837 pKa = 11.84DD838 pKa = 3.79NPFSGCQNGIAIQIGRR854 pKa = 11.84NALSSSGTAIIHH866 pKa = 5.78NNIISGYY873 pKa = 7.77QKK875 pKa = 10.87GAIVVDD881 pKa = 3.8NSGSNATITNNTITGTGTTDD901 pKa = 2.72VTAQNGIQVSRR912 pKa = 11.84GATATLGGNTISGNSFHH929 pKa = 7.46EE930 pKa = 5.31EE931 pKa = 3.81GSQWDD936 pKa = 3.41WGACGILLYY945 pKa = 10.71QSGAVSLTGGNNISGNDD962 pKa = 3.51QNYY965 pKa = 8.75YY966 pKa = 10.84AYY968 pKa = 10.34DD969 pKa = 3.41VAGNLILGAEE979 pKa = 4.3TFGASTAPVTFGYY992 pKa = 10.78NIADD996 pKa = 3.69YY997 pKa = 10.44TNQNIDD1003 pKa = 3.07ASQCTFAEE1011 pKa = 4.66GNPASAVLSEE1021 pKa = 4.74LFDD1024 pKa = 4.48IEE1026 pKa = 4.36DD1027 pKa = 4.87RR1028 pKa = 11.84IWHH1031 pKa = 5.99SVDD1034 pKa = 3.3DD1035 pKa = 3.92QTISGFVNIKK1045 pKa = 10.04AGNVYY1050 pKa = 8.47VTHH1053 pKa = 6.77TEE1055 pKa = 3.91SGAKK1059 pKa = 8.36IQYY1062 pKa = 10.2GIDD1065 pKa = 3.32AASVNDD1071 pKa = 3.87VIHH1074 pKa = 6.25VKK1076 pKa = 10.5EE1077 pKa = 4.19GTYY1080 pKa = 10.35SEE1082 pKa = 5.19NITVDD1087 pKa = 3.08KK1088 pKa = 11.09DD1089 pKa = 3.64VTLLGANAGVACDD1102 pKa = 3.64TRR1104 pKa = 11.84GAEE1107 pKa = 4.09STIAGTGGAAVSVASDD1123 pKa = 3.41GVTIDD1128 pKa = 3.61GFEE1131 pKa = 4.01ITNPTGNYY1139 pKa = 9.39AVNSTGNNNLLLQYY1153 pKa = 11.37NNVNDD1158 pKa = 3.82VGTSGTPGNVHH1169 pKa = 6.66AVAISMGSANTDD1181 pKa = 3.16DD1182 pKa = 5.51VKK1184 pKa = 10.82IQHH1187 pKa = 6.1NCISNINNAGNTHH1200 pKa = 6.35SGSGIAVGFSTATTDD1215 pKa = 3.11ITNLYY1220 pKa = 9.24IGYY1223 pKa = 8.29NTISQVNSNNALDD1236 pKa = 3.91YY1237 pKa = 11.25ASGGRR1242 pKa = 11.84GANGIILNVGANSTGKK1258 pKa = 8.41VTDD1261 pKa = 3.97GVVEE1265 pKa = 4.23YY1266 pKa = 9.08NTIHH1270 pKa = 7.15DD1271 pKa = 4.02LSGYY1275 pKa = 7.27WVHH1278 pKa = 6.89GVGLEE1283 pKa = 4.14GNTPGALVQNNLIYY1297 pKa = 10.88NLVSTYY1303 pKa = 10.37TDD1305 pKa = 2.93IPGVGVKK1312 pKa = 10.3VEE1314 pKa = 4.74DD1315 pKa = 3.5NTGAATVSVNNNSFTSLSPGILNAVSGATVNATCNWFAADD1355 pKa = 3.69VTEE1358 pKa = 4.87KK1359 pKa = 11.01SLGDD1363 pKa = 3.5VTVVPWLVDD1372 pKa = 3.31GTNSVASGPGFEE1384 pKa = 4.71PATACVACAISIDD1397 pKa = 3.64AVVTNVSCPGGADD1410 pKa = 3.49DD1411 pKa = 5.16GAIDD1415 pKa = 3.63VTVTNGVSPYY1425 pKa = 8.55TYY1427 pKa = 10.48SWTGPDD1433 pKa = 4.07GFTSPDD1439 pKa = 3.24EE1440 pKa = 5.24DD1441 pKa = 3.76ISGLAAGTYY1450 pKa = 9.97NLLVTADD1457 pKa = 3.82NGCSKK1462 pKa = 10.72EE1463 pKa = 4.0KK1464 pKa = 10.65VVVVGTDD1471 pKa = 3.19ADD1473 pKa = 3.95VTAPTITEE1481 pKa = 4.02ALDD1484 pKa = 3.21VLTEE1488 pKa = 4.13EE1489 pKa = 5.15GCTDD1493 pKa = 3.69ADD1495 pKa = 3.81VPVAKK1500 pKa = 8.37TTVAEE1505 pKa = 4.16LEE1507 pKa = 4.2AMGVIIDD1514 pKa = 5.1DD1515 pKa = 4.0CTLDD1519 pKa = 3.44EE1520 pKa = 4.8NMTVSYY1526 pKa = 10.93SDD1528 pKa = 4.14ASAGTCPVVVTRR1540 pKa = 11.84TYY1542 pKa = 10.86TVTDD1546 pKa = 3.91ASNNSVTVDD1555 pKa = 3.1QTINIEE1561 pKa = 4.38DD1562 pKa = 3.77NTPPSIACPPPIAVKK1577 pKa = 10.4MNDD1580 pKa = 2.96GCEE1583 pKa = 3.83YY1584 pKa = 11.03DD1585 pKa = 3.63PAIGSGIGVATATDD1599 pKa = 3.62NCSDD1603 pKa = 3.87AVDD1606 pKa = 3.55IQISNDD1612 pKa = 2.49ITGNLTEE1619 pKa = 4.8GEE1621 pKa = 4.26NFITWTAVDD1630 pKa = 3.66EE1631 pKa = 4.74CNNSVTCTQKK1641 pKa = 9.23VTVIRR1646 pKa = 11.84NTLSGTVMYY1655 pKa = 10.62NNTAQTPMANVVLKK1669 pKa = 11.06LFDD1672 pKa = 4.61SNDD1675 pKa = 3.6DD1676 pKa = 3.59QVGDD1680 pKa = 3.98DD1681 pKa = 3.8VVTSEE1686 pKa = 4.35TNPGNFEE1693 pKa = 4.06FTGLCAGDD1701 pKa = 3.8YY1702 pKa = 9.97TVVASDD1708 pKa = 3.56TSKK1711 pKa = 10.92VGYY1714 pKa = 10.31INATDD1719 pKa = 3.64AGAANAWGTAGGDD1732 pKa = 3.11IEE1734 pKa = 4.88YY1735 pKa = 10.69VQFLAGDD1742 pKa = 3.66VNDD1745 pKa = 4.07NLLINSTDD1753 pKa = 3.09AQAIQNYY1760 pKa = 8.26FVFGGSFTKK1769 pKa = 10.04PWEE1772 pKa = 3.9YY1773 pKa = 10.25WSAGSVIHH1781 pKa = 6.85SNYY1784 pKa = 10.33NPFSSNAIPPHH1795 pKa = 5.91PAAIEE1800 pKa = 4.13VVISGGDD1807 pKa = 3.51VTHH1810 pKa = 7.5DD1811 pKa = 4.84LYY1813 pKa = 11.59AQATGDD1819 pKa = 3.8FNGSFGPTLKK1829 pKa = 10.73SAGSSVRR1836 pKa = 11.84LSADD1840 pKa = 2.77RR1841 pKa = 11.84NMNVEE1846 pKa = 4.15EE1847 pKa = 4.06NQVFNLPLRR1856 pKa = 11.84SEE1858 pKa = 4.37FAMEE1862 pKa = 4.04VGAVSMILDD1871 pKa = 3.62MPEE1874 pKa = 4.81DD1875 pKa = 4.05VVQVTGVQIAGSEE1888 pKa = 4.49VPVTWAMKK1896 pKa = 10.45DD1897 pKa = 3.13NEE1899 pKa = 5.07LRR1901 pKa = 11.84IGWNSLNPVYY1911 pKa = 10.75VGEE1914 pKa = 4.17NDD1916 pKa = 3.26AFVVLKK1922 pKa = 10.47LQTSANFTEE1931 pKa = 4.62GQLMDD1936 pKa = 3.85IGLVYY1941 pKa = 11.02NPLNEE1946 pKa = 4.13LADD1949 pKa = 4.07GNFEE1953 pKa = 4.76PIEE1956 pKa = 4.1NAALKK1961 pKa = 9.53VARR1964 pKa = 11.84VGNGLTDD1971 pKa = 3.22INEE1974 pKa = 4.59DD1975 pKa = 3.46LDD1977 pKa = 3.75IDD1979 pKa = 4.35KK1980 pKa = 8.13MTFSNYY1986 pKa = 9.15PNPFSEE1992 pKa = 4.56STTLMYY1998 pKa = 8.58TIPVDD2003 pKa = 3.5GKK2005 pKa = 11.33VNISVYY2011 pKa = 10.38NALGQLVTTIVDD2023 pKa = 3.35EE2024 pKa = 4.23NQRR2027 pKa = 11.84TGRR2030 pKa = 11.84YY2031 pKa = 7.14TINNCGDD2038 pKa = 3.47EE2039 pKa = 4.74LQPGMYY2045 pKa = 9.15ISKK2048 pKa = 10.67LMLTNGTEE2056 pKa = 4.41SMVSTIKK2063 pKa = 11.02LNVVKK2068 pKa = 10.82
Molecular weight: 214.89 kDa
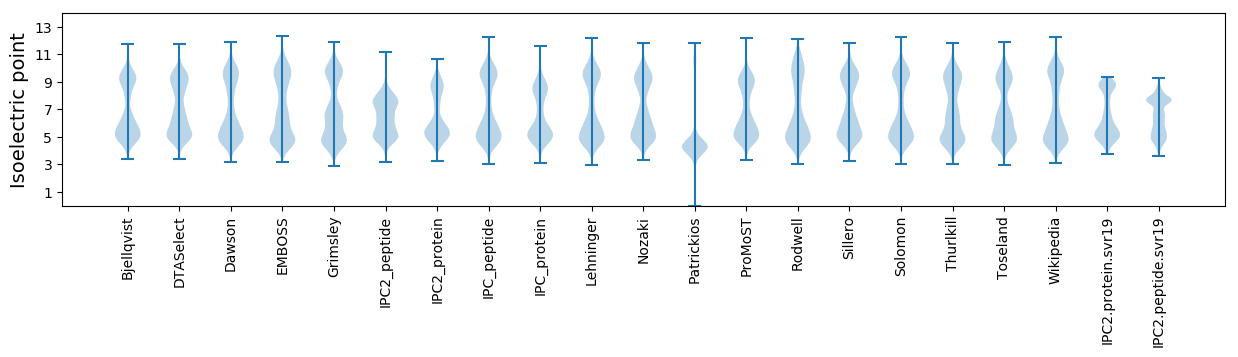
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1M6IEJ3|A0A1M6IEJ3_9BACT Uncharacterized protein OS=Tangfeifania diversioriginum OX=1168035 GN=SAMN05444280_11675 PE=4 SV=1
MM1 pKa = 7.9IITTQQPASSAPRR14 pKa = 11.84KK15 pKa = 7.95KK16 pKa = 8.85TRR18 pKa = 11.84RR19 pKa = 11.84CLCYY23 pKa = 10.37NIRR26 pKa = 11.84EE27 pKa = 4.16NLDD30 pKa = 3.24EE31 pKa = 4.52KK32 pKa = 10.17MKK34 pKa = 10.64EE35 pKa = 3.93MRR37 pKa = 11.84LKK39 pKa = 10.35RR40 pKa = 11.84AIIISAFQADD50 pKa = 4.14YY51 pKa = 11.54QMMVGNVSGPAPGGEE66 pKa = 3.77FRR68 pKa = 11.84AFSPVMLWAMPIVLLSKK85 pKa = 10.6AFSLLGYY92 pKa = 9.22RR93 pKa = 11.84AVYY96 pKa = 9.58LVRR99 pKa = 11.84VLSISLQAEE108 pKa = 3.89SLTYY112 pKa = 10.5HH113 pKa = 6.44NLGHH117 pKa = 6.96RR118 pKa = 11.84PRR120 pKa = 11.84PGNHH124 pKa = 5.89SRR126 pKa = 11.84PDD128 pKa = 3.56PYY130 pKa = 11.47LMSTHH135 pKa = 7.2LYY137 pKa = 7.48MAARR141 pKa = 11.84SFMTVHH147 pKa = 7.18RR148 pKa = 11.84PRR150 pKa = 11.84LSATRR155 pKa = 11.84SKK157 pKa = 9.83RR158 pKa = 11.84TAARR162 pKa = 11.84SACRR166 pKa = 11.84SGSEE170 pKa = 4.18HH171 pKa = 7.28LPISNYY177 pKa = 10.06HH178 pKa = 5.47YY179 pKa = 10.2GYY181 pKa = 9.65PAMLQKK187 pKa = 10.51ISS189 pKa = 3.31
MM1 pKa = 7.9IITTQQPASSAPRR14 pKa = 11.84KK15 pKa = 7.95KK16 pKa = 8.85TRR18 pKa = 11.84RR19 pKa = 11.84CLCYY23 pKa = 10.37NIRR26 pKa = 11.84EE27 pKa = 4.16NLDD30 pKa = 3.24EE31 pKa = 4.52KK32 pKa = 10.17MKK34 pKa = 10.64EE35 pKa = 3.93MRR37 pKa = 11.84LKK39 pKa = 10.35RR40 pKa = 11.84AIIISAFQADD50 pKa = 4.14YY51 pKa = 11.54QMMVGNVSGPAPGGEE66 pKa = 3.77FRR68 pKa = 11.84AFSPVMLWAMPIVLLSKK85 pKa = 10.6AFSLLGYY92 pKa = 9.22RR93 pKa = 11.84AVYY96 pKa = 9.58LVRR99 pKa = 11.84VLSISLQAEE108 pKa = 3.89SLTYY112 pKa = 10.5HH113 pKa = 6.44NLGHH117 pKa = 6.96RR118 pKa = 11.84PRR120 pKa = 11.84PGNHH124 pKa = 5.89SRR126 pKa = 11.84PDD128 pKa = 3.56PYY130 pKa = 11.47LMSTHH135 pKa = 7.2LYY137 pKa = 7.48MAARR141 pKa = 11.84SFMTVHH147 pKa = 7.18RR148 pKa = 11.84PRR150 pKa = 11.84LSATRR155 pKa = 11.84SKK157 pKa = 9.83RR158 pKa = 11.84TAARR162 pKa = 11.84SACRR166 pKa = 11.84SGSEE170 pKa = 4.18HH171 pKa = 7.28LPISNYY177 pKa = 10.06HH178 pKa = 5.47YY179 pKa = 10.2GYY181 pKa = 9.65PAMLQKK187 pKa = 10.51ISS189 pKa = 3.31
Molecular weight: 21.42 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1425793 |
26 |
3828 |
347.5 |
39.32 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.376 ± 0.032 | 0.764 ± 0.012 |
5.355 ± 0.033 | 7.41 ± 0.035 |
5.371 ± 0.029 | 6.765 ± 0.036 |
1.828 ± 0.018 | 7.298 ± 0.033 |
7.138 ± 0.049 | 9.105 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.327 ± 0.017 | 5.848 ± 0.031 |
3.836 ± 0.02 | 3.433 ± 0.022 |
4.015 ± 0.029 | 6.199 ± 0.033 |
5.351 ± 0.04 | 6.347 ± 0.03 |
1.309 ± 0.018 | 3.926 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
