
Micromonospora sp. 4G51
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micromonosporales; Micromonosporaceae; Micromonospora; unclassified Micromonospora
Average proteome isoelectric point is 6.69
Get precalculated fractions of proteins
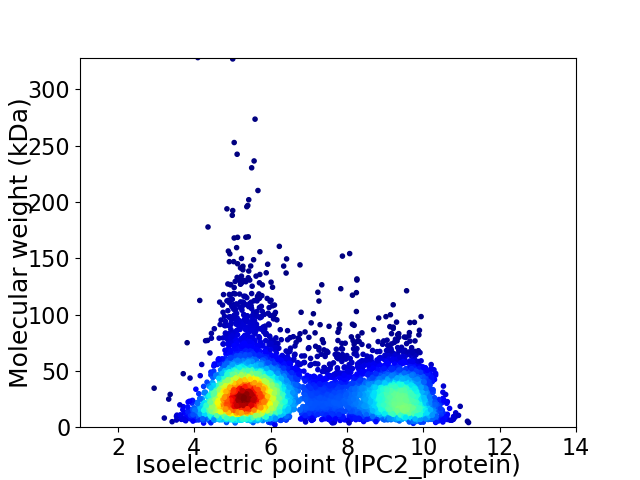
Virtual 2D-PAGE plot for 6887 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A317DN82|A0A317DN82_9ACTN Uncharacterized protein (Fragment) OS=Micromonospora sp. 4G51 OX=2202420 GN=DKT69_10200 PE=4 SV=1
MM1 pKa = 6.97TALAFTVSAPAQANDD16 pKa = 3.58HH17 pKa = 6.55EE18 pKa = 4.66NLPGEE23 pKa = 4.2PTITLPINDD32 pKa = 3.78SAAGGTYY39 pKa = 10.09NQDD42 pKa = 3.11FTRR45 pKa = 11.84VYY47 pKa = 11.41ANTTPDD53 pKa = 3.31GTPVYY58 pKa = 10.08IYY60 pKa = 10.52VPKK63 pKa = 9.73GTPLDD68 pKa = 3.67GTAPAGVASLDD79 pKa = 3.44PQFDD83 pKa = 4.12HH84 pKa = 7.02NPADD88 pKa = 3.74GFTPCATATAAQFTLTQAEE107 pKa = 4.2INYY110 pKa = 10.07VGDD113 pKa = 3.67KK114 pKa = 10.85LADD117 pKa = 3.45QIVAVDD123 pKa = 3.83EE124 pKa = 4.23EE125 pKa = 5.47HH126 pKa = 6.64YY127 pKa = 11.36GPMDD131 pKa = 3.9AADD134 pKa = 3.98PAEE137 pKa = 4.4PASDD141 pKa = 3.6SLVMVLYY148 pKa = 10.64NIQDD152 pKa = 3.5DD153 pKa = 4.51AYY155 pKa = 9.53YY156 pKa = 10.96DD157 pKa = 3.96CAQTSYY163 pKa = 9.62TAGYY167 pKa = 7.63FAPDD171 pKa = 4.9FIDD174 pKa = 3.53SVGMNVIVVDD184 pKa = 3.98ALDD187 pKa = 3.24WANRR191 pKa = 11.84VGPNDD196 pKa = 4.06SPWRR200 pKa = 11.84DD201 pKa = 3.26ANPANDD207 pKa = 3.77RR208 pKa = 11.84PEE210 pKa = 4.76LYY212 pKa = 9.95EE213 pKa = 3.8GTVAHH218 pKa = 6.86EE219 pKa = 4.26LQHH222 pKa = 6.7LLHH225 pKa = 6.77NYY227 pKa = 9.69SDD229 pKa = 4.0PGEE232 pKa = 4.2LSWVDD237 pKa = 3.2EE238 pKa = 4.25GLADD242 pKa = 4.89FAIFLNGLDD251 pKa = 3.78AGGSHH256 pKa = 7.78LSNQQVFYY264 pKa = 11.03EE265 pKa = 4.46EE266 pKa = 4.2TSLTRR271 pKa = 11.84WGGTLANYY279 pKa = 7.71GASFTYY285 pKa = 9.52FQYY288 pKa = 11.02LWEE291 pKa = 4.13QAGGNGDD298 pKa = 3.71GTYY301 pKa = 10.08TPDD304 pKa = 3.62KK305 pKa = 10.99QYY307 pKa = 11.45DD308 pKa = 3.93DD309 pKa = 3.9KK310 pKa = 11.99AGDD313 pKa = 4.09LLIKK317 pKa = 9.82TIFEE321 pKa = 4.07EE322 pKa = 4.52QADD325 pKa = 3.96GMTGVQNAIDD335 pKa = 4.61DD336 pKa = 4.32FNAATGSHH344 pKa = 6.09LRR346 pKa = 11.84SAADD350 pKa = 4.09LFRR353 pKa = 11.84DD354 pKa = 3.46WSVAVYY360 pKa = 10.72LDD362 pKa = 4.64DD363 pKa = 5.03EE364 pKa = 4.56NSPTYY369 pKa = 10.08DD370 pKa = 2.4IKK372 pKa = 11.3AFNFGDD378 pKa = 3.91PADD381 pKa = 4.16TAWTIDD387 pKa = 3.2IADD390 pKa = 3.78DD391 pKa = 4.0VFWSGRR397 pKa = 11.84GSYY400 pKa = 9.81QGATPEE406 pKa = 4.3AKK408 pKa = 9.29WARR411 pKa = 11.84LKK413 pKa = 10.72NRR415 pKa = 11.84PDD417 pKa = 3.11TTAVPFGMSVEE428 pKa = 4.27RR429 pKa = 11.84FRR431 pKa = 11.84NPGPTVAVAFDD442 pKa = 4.51GDD444 pKa = 4.4DD445 pKa = 3.37QTVIAPHH452 pKa = 6.61TGTTHH457 pKa = 6.59WYY459 pKa = 10.12AGYY462 pKa = 10.32EE463 pKa = 4.16SQSDD467 pKa = 3.84NILNVDD473 pKa = 3.34TAGPVSSLDD482 pKa = 3.27FWSWNFIEE490 pKa = 5.16EE491 pKa = 4.13GWDD494 pKa = 3.28YY495 pKa = 11.62GFAEE499 pKa = 4.64ALVGGDD505 pKa = 2.98WVTVPLRR512 pKa = 11.84NDD514 pKa = 3.02AGQVVTTNDD523 pKa = 3.47NPHH526 pKa = 6.32GNNTEE531 pKa = 4.3GNGLTGTSGGEE542 pKa = 4.02YY543 pKa = 10.35FVDD546 pKa = 3.79DD547 pKa = 4.16PVYY550 pKa = 11.1VHH552 pKa = 6.54LTANLPAGATDD563 pKa = 3.03VRR565 pKa = 11.84FRR567 pKa = 11.84YY568 pKa = 8.58STDD571 pKa = 2.83AAYY574 pKa = 10.92LDD576 pKa = 4.21TGWFVDD582 pKa = 3.9DD583 pKa = 4.01VRR585 pKa = 11.84VNGADD590 pKa = 3.3ATLSSPAGDD599 pKa = 3.13WFSTTGTQDD608 pKa = 3.32NNWTVQVVSHH618 pKa = 7.15CDD620 pKa = 3.41LTPGTTSAGEE630 pKa = 4.06ITDD633 pKa = 4.08GAGNWVYY640 pKa = 11.01RR641 pKa = 11.84FTGDD645 pKa = 2.7QFTRR649 pKa = 11.84SGLQTKK655 pKa = 10.03CAGGNQDD662 pKa = 4.97DD663 pKa = 4.27FAVLISNLPTGDD675 pKa = 3.92LTFLDD680 pKa = 3.43ADD682 pKa = 3.68YY683 pKa = 10.41TFRR686 pKa = 11.84VTNTGNKK693 pKa = 9.04KK694 pKa = 10.25
MM1 pKa = 6.97TALAFTVSAPAQANDD16 pKa = 3.58HH17 pKa = 6.55EE18 pKa = 4.66NLPGEE23 pKa = 4.2PTITLPINDD32 pKa = 3.78SAAGGTYY39 pKa = 10.09NQDD42 pKa = 3.11FTRR45 pKa = 11.84VYY47 pKa = 11.41ANTTPDD53 pKa = 3.31GTPVYY58 pKa = 10.08IYY60 pKa = 10.52VPKK63 pKa = 9.73GTPLDD68 pKa = 3.67GTAPAGVASLDD79 pKa = 3.44PQFDD83 pKa = 4.12HH84 pKa = 7.02NPADD88 pKa = 3.74GFTPCATATAAQFTLTQAEE107 pKa = 4.2INYY110 pKa = 10.07VGDD113 pKa = 3.67KK114 pKa = 10.85LADD117 pKa = 3.45QIVAVDD123 pKa = 3.83EE124 pKa = 4.23EE125 pKa = 5.47HH126 pKa = 6.64YY127 pKa = 11.36GPMDD131 pKa = 3.9AADD134 pKa = 3.98PAEE137 pKa = 4.4PASDD141 pKa = 3.6SLVMVLYY148 pKa = 10.64NIQDD152 pKa = 3.5DD153 pKa = 4.51AYY155 pKa = 9.53YY156 pKa = 10.96DD157 pKa = 3.96CAQTSYY163 pKa = 9.62TAGYY167 pKa = 7.63FAPDD171 pKa = 4.9FIDD174 pKa = 3.53SVGMNVIVVDD184 pKa = 3.98ALDD187 pKa = 3.24WANRR191 pKa = 11.84VGPNDD196 pKa = 4.06SPWRR200 pKa = 11.84DD201 pKa = 3.26ANPANDD207 pKa = 3.77RR208 pKa = 11.84PEE210 pKa = 4.76LYY212 pKa = 9.95EE213 pKa = 3.8GTVAHH218 pKa = 6.86EE219 pKa = 4.26LQHH222 pKa = 6.7LLHH225 pKa = 6.77NYY227 pKa = 9.69SDD229 pKa = 4.0PGEE232 pKa = 4.2LSWVDD237 pKa = 3.2EE238 pKa = 4.25GLADD242 pKa = 4.89FAIFLNGLDD251 pKa = 3.78AGGSHH256 pKa = 7.78LSNQQVFYY264 pKa = 11.03EE265 pKa = 4.46EE266 pKa = 4.2TSLTRR271 pKa = 11.84WGGTLANYY279 pKa = 7.71GASFTYY285 pKa = 9.52FQYY288 pKa = 11.02LWEE291 pKa = 4.13QAGGNGDD298 pKa = 3.71GTYY301 pKa = 10.08TPDD304 pKa = 3.62KK305 pKa = 10.99QYY307 pKa = 11.45DD308 pKa = 3.93DD309 pKa = 3.9KK310 pKa = 11.99AGDD313 pKa = 4.09LLIKK317 pKa = 9.82TIFEE321 pKa = 4.07EE322 pKa = 4.52QADD325 pKa = 3.96GMTGVQNAIDD335 pKa = 4.61DD336 pKa = 4.32FNAATGSHH344 pKa = 6.09LRR346 pKa = 11.84SAADD350 pKa = 4.09LFRR353 pKa = 11.84DD354 pKa = 3.46WSVAVYY360 pKa = 10.72LDD362 pKa = 4.64DD363 pKa = 5.03EE364 pKa = 4.56NSPTYY369 pKa = 10.08DD370 pKa = 2.4IKK372 pKa = 11.3AFNFGDD378 pKa = 3.91PADD381 pKa = 4.16TAWTIDD387 pKa = 3.2IADD390 pKa = 3.78DD391 pKa = 4.0VFWSGRR397 pKa = 11.84GSYY400 pKa = 9.81QGATPEE406 pKa = 4.3AKK408 pKa = 9.29WARR411 pKa = 11.84LKK413 pKa = 10.72NRR415 pKa = 11.84PDD417 pKa = 3.11TTAVPFGMSVEE428 pKa = 4.27RR429 pKa = 11.84FRR431 pKa = 11.84NPGPTVAVAFDD442 pKa = 4.51GDD444 pKa = 4.4DD445 pKa = 3.37QTVIAPHH452 pKa = 6.61TGTTHH457 pKa = 6.59WYY459 pKa = 10.12AGYY462 pKa = 10.32EE463 pKa = 4.16SQSDD467 pKa = 3.84NILNVDD473 pKa = 3.34TAGPVSSLDD482 pKa = 3.27FWSWNFIEE490 pKa = 5.16EE491 pKa = 4.13GWDD494 pKa = 3.28YY495 pKa = 11.62GFAEE499 pKa = 4.64ALVGGDD505 pKa = 2.98WVTVPLRR512 pKa = 11.84NDD514 pKa = 3.02AGQVVTTNDD523 pKa = 3.47NPHH526 pKa = 6.32GNNTEE531 pKa = 4.3GNGLTGTSGGEE542 pKa = 4.02YY543 pKa = 10.35FVDD546 pKa = 3.79DD547 pKa = 4.16PVYY550 pKa = 11.1VHH552 pKa = 6.54LTANLPAGATDD563 pKa = 3.03VRR565 pKa = 11.84FRR567 pKa = 11.84YY568 pKa = 8.58STDD571 pKa = 2.83AAYY574 pKa = 10.92LDD576 pKa = 4.21TGWFVDD582 pKa = 3.9DD583 pKa = 4.01VRR585 pKa = 11.84VNGADD590 pKa = 3.3ATLSSPAGDD599 pKa = 3.13WFSTTGTQDD608 pKa = 3.32NNWTVQVVSHH618 pKa = 7.15CDD620 pKa = 3.41LTPGTTSAGEE630 pKa = 4.06ITDD633 pKa = 4.08GAGNWVYY640 pKa = 11.01RR641 pKa = 11.84FTGDD645 pKa = 2.7QFTRR649 pKa = 11.84SGLQTKK655 pKa = 10.03CAGGNQDD662 pKa = 4.97DD663 pKa = 4.27FAVLISNLPTGDD675 pKa = 3.92LTFLDD680 pKa = 3.43ADD682 pKa = 3.68YY683 pKa = 10.41TFRR686 pKa = 11.84VTNTGNKK693 pKa = 9.04KK694 pKa = 10.25
Molecular weight: 75.03 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A317DJ32|A0A317DJ32_9ACTN Aquaporin OS=Micromonospora sp. 4G51 OX=2202420 GN=DKT69_14625 PE=3 SV=1
MM1 pKa = 7.28GSVVKK6 pKa = 10.49KK7 pKa = 9.45RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.48RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.7LLRR22 pKa = 11.84KK23 pKa = 7.84TRR25 pKa = 11.84VQRR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84LGKK33 pKa = 10.04
MM1 pKa = 7.28GSVVKK6 pKa = 10.49KK7 pKa = 9.45RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.48RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.7LLRR22 pKa = 11.84KK23 pKa = 7.84TRR25 pKa = 11.84VQRR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84LGKK33 pKa = 10.04
Molecular weight: 4.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2147859 |
21 |
3225 |
311.9 |
33.42 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.97 ± 0.044 | 0.761 ± 0.009 |
6.058 ± 0.023 | 5.056 ± 0.028 |
2.683 ± 0.017 | 9.294 ± 0.032 |
2.177 ± 0.017 | 3.2 ± 0.02 |
1.804 ± 0.022 | 10.528 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.608 ± 0.013 | 1.824 ± 0.021 |
6.269 ± 0.027 | 2.812 ± 0.018 |
8.564 ± 0.041 | 4.643 ± 0.023 |
6.059 ± 0.03 | 8.941 ± 0.03 |
1.651 ± 0.013 | 2.098 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
