
[Clostridium] innocuum 2959
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Erysipelotrichia; Erysipelotrichales; Erysipelotrichaceae; Erysipelatoclostridium; [Clostridium] innocuum
Average proteome isoelectric point is 6.22
Get precalculated fractions of proteins
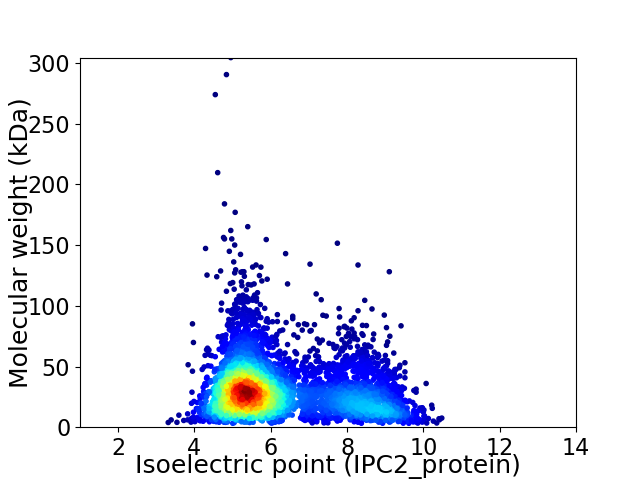
Virtual 2D-PAGE plot for 4653 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|N9WUR9|N9WUR9_CLOIN HTH deoR-type domain-containing protein OS=[Clostridium] innocuum 2959 OX=999413 GN=HMPREF1094_02081 PE=4 SV=1
MM1 pKa = 7.66HH2 pKa = 7.31YY3 pKa = 10.74DD4 pKa = 5.22DD5 pKa = 6.35YY6 pKa = 12.1DD7 pKa = 4.83FDD9 pKa = 6.32DD10 pKa = 4.77KK11 pKa = 11.75NCCNEE16 pKa = 4.3CSSSSCDD23 pKa = 3.13QGDD26 pKa = 3.86YY27 pKa = 10.96DD28 pKa = 4.41VCCCPGPRR36 pKa = 11.84GPRR39 pKa = 11.84GFRR42 pKa = 11.84GPAGEE47 pKa = 4.6AGATGATGATGNTGLTGATGATGLSEE73 pKa = 4.58TITIRR78 pKa = 11.84STTTADD84 pKa = 3.44PGTPAQVHH92 pKa = 6.7DD93 pKa = 4.31SGGPNHH99 pKa = 6.2VLDD102 pKa = 4.39FVIPRR107 pKa = 11.84GDD109 pKa = 3.37TGVTGATGPTGPGVGATGPTGATGATGNTGATGPIGATGPTGANGNTGPTGAIGATGEE167 pKa = 4.14NGATGPTGPAGPTGATGSTGAAGATGVTGPTGDD200 pKa = 3.82IGPTGPTGATGSTGAAGATGVTGATGDD227 pKa = 3.57IGPTGPTGATGPTGATGNTGATGTTGVTGADD258 pKa = 3.76GAIGPTGPIGDD269 pKa = 5.16PGPQGEE275 pKa = 5.01PGPQGEE281 pKa = 4.92PGPQGEE287 pKa = 4.45QGPQGEE293 pKa = 4.62TGATPIVRR301 pKa = 11.84VGSTTTSEE309 pKa = 4.34PGTPADD315 pKa = 3.32VSYY318 pKa = 11.62VEE320 pKa = 4.37TPDD323 pKa = 4.73GIEE326 pKa = 4.17LNFIIPRR333 pKa = 11.84GDD335 pKa = 3.22TGPGGGGGGLLAYY348 pKa = 9.67GGKK351 pKa = 10.59YY352 pKa = 9.69NDD354 pKa = 3.8SAQTLNLLIGAEE366 pKa = 4.09QQLPLPVDD374 pKa = 3.81MSASNVDD381 pKa = 4.21LAPVNALTIVTSGVYY396 pKa = 9.83EE397 pKa = 3.84INYY400 pKa = 7.27MFNASASLGAAVTLAVRR417 pKa = 11.84RR418 pKa = 11.84NGAAIPSTEE427 pKa = 3.91EE428 pKa = 3.44RR429 pKa = 11.84HH430 pKa = 5.85LLAIATEE437 pKa = 4.59SIYY440 pKa = 10.68SGSVIEE446 pKa = 4.77PLSAGDD452 pKa = 4.17IIDD455 pKa = 3.87MAVSAAIALTLTLSTGVTVTLSVKK479 pKa = 10.35RR480 pKa = 11.84LNDD483 pKa = 3.35ALANN487 pKa = 3.9
MM1 pKa = 7.66HH2 pKa = 7.31YY3 pKa = 10.74DD4 pKa = 5.22DD5 pKa = 6.35YY6 pKa = 12.1DD7 pKa = 4.83FDD9 pKa = 6.32DD10 pKa = 4.77KK11 pKa = 11.75NCCNEE16 pKa = 4.3CSSSSCDD23 pKa = 3.13QGDD26 pKa = 3.86YY27 pKa = 10.96DD28 pKa = 4.41VCCCPGPRR36 pKa = 11.84GPRR39 pKa = 11.84GFRR42 pKa = 11.84GPAGEE47 pKa = 4.6AGATGATGATGNTGLTGATGATGLSEE73 pKa = 4.58TITIRR78 pKa = 11.84STTTADD84 pKa = 3.44PGTPAQVHH92 pKa = 6.7DD93 pKa = 4.31SGGPNHH99 pKa = 6.2VLDD102 pKa = 4.39FVIPRR107 pKa = 11.84GDD109 pKa = 3.37TGVTGATGPTGPGVGATGPTGATGATGNTGATGPIGATGPTGANGNTGPTGAIGATGEE167 pKa = 4.14NGATGPTGPAGPTGATGSTGAAGATGVTGPTGDD200 pKa = 3.82IGPTGPTGATGSTGAAGATGVTGATGDD227 pKa = 3.57IGPTGPTGATGPTGATGNTGATGTTGVTGADD258 pKa = 3.76GAIGPTGPIGDD269 pKa = 5.16PGPQGEE275 pKa = 5.01PGPQGEE281 pKa = 4.92PGPQGEE287 pKa = 4.45QGPQGEE293 pKa = 4.62TGATPIVRR301 pKa = 11.84VGSTTTSEE309 pKa = 4.34PGTPADD315 pKa = 3.32VSYY318 pKa = 11.62VEE320 pKa = 4.37TPDD323 pKa = 4.73GIEE326 pKa = 4.17LNFIIPRR333 pKa = 11.84GDD335 pKa = 3.22TGPGGGGGGLLAYY348 pKa = 9.67GGKK351 pKa = 10.59YY352 pKa = 9.69NDD354 pKa = 3.8SAQTLNLLIGAEE366 pKa = 4.09QQLPLPVDD374 pKa = 3.81MSASNVDD381 pKa = 4.21LAPVNALTIVTSGVYY396 pKa = 9.83EE397 pKa = 3.84INYY400 pKa = 7.27MFNASASLGAAVTLAVRR417 pKa = 11.84RR418 pKa = 11.84NGAAIPSTEE427 pKa = 3.91EE428 pKa = 3.44RR429 pKa = 11.84HH430 pKa = 5.85LLAIATEE437 pKa = 4.59SIYY440 pKa = 10.68SGSVIEE446 pKa = 4.77PLSAGDD452 pKa = 4.17IIDD455 pKa = 3.87MAVSAAIALTLTLSTGVTVTLSVKK479 pKa = 10.35RR480 pKa = 11.84LNDD483 pKa = 3.35ALANN487 pKa = 3.9
Molecular weight: 46.14 kDa
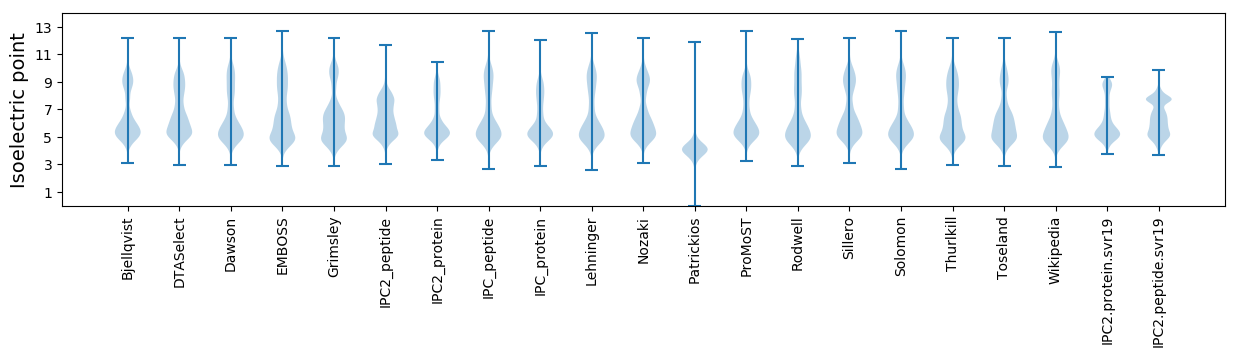
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|N9VCF8|N9VCF8_CLOIN Peptide chain release factor 1 OS=[Clostridium] innocuum 2959 OX=999413 GN=prfA PE=3 SV=1
MM1 pKa = 7.41FFHH4 pKa = 6.36SWRR7 pKa = 11.84RR8 pKa = 11.84KK9 pKa = 8.67RR10 pKa = 11.84KK11 pKa = 8.75LKK13 pKa = 10.22RR14 pKa = 11.84FRR16 pKa = 11.84PIMTLHH22 pKa = 7.11LNRR25 pKa = 11.84TLHH28 pKa = 6.71HH29 pKa = 6.42EE30 pKa = 4.2FHH32 pKa = 6.56PTFLKK37 pKa = 10.84AKK39 pKa = 8.0TVLRR43 pKa = 11.84EE44 pKa = 3.85VRR46 pKa = 11.84SSVIRR51 pKa = 11.84KK52 pKa = 9.06EE53 pKa = 3.92EE54 pKa = 4.03HH55 pKa = 4.86THH57 pKa = 6.6HH58 pKa = 6.43IHH60 pKa = 7.73HH61 pKa = 6.71MMTLHH66 pKa = 5.69VHH68 pKa = 5.11QAQTFLMLNHH78 pKa = 6.12QNDD81 pKa = 3.69EE82 pKa = 4.08RR83 pKa = 11.84SLIIQSKK90 pKa = 9.78KK91 pKa = 10.05VINRR95 pKa = 11.84KK96 pKa = 8.59YY97 pKa = 9.95PSRR100 pKa = 11.84ILQLHH105 pKa = 6.41LAKK108 pKa = 10.53QSIEE112 pKa = 3.89RR113 pKa = 11.84LAEE116 pKa = 3.77QRR118 pKa = 11.84LLEE121 pKa = 4.3EE122 pKa = 3.72KK123 pKa = 10.25SYY125 pKa = 11.54YY126 pKa = 10.01KK127 pKa = 9.88YY128 pKa = 10.68HH129 pKa = 6.49PAKK132 pKa = 10.44AKK134 pKa = 9.84EE135 pKa = 3.81LAVSYY140 pKa = 9.22LHH142 pKa = 6.56NRR144 pKa = 11.84EE145 pKa = 4.17IKK147 pKa = 10.27NCLNTHH153 pKa = 5.9MNRR156 pKa = 11.84LSQYY160 pKa = 10.07SYY162 pKa = 10.89AGSLFRR168 pKa = 11.84SSLFTTLSSQSWKK181 pKa = 10.11QVLIPVIYY189 pKa = 8.79QQSRR193 pKa = 11.84ALIHH197 pKa = 6.43RR198 pKa = 11.84EE199 pKa = 3.63KK200 pKa = 10.52IQLQRR205 pKa = 11.84EE206 pKa = 4.52VISEE210 pKa = 4.02KK211 pKa = 10.83SRR213 pKa = 11.84DD214 pKa = 3.59EE215 pKa = 3.79MHH217 pKa = 6.56VEE219 pKa = 4.06RR220 pKa = 11.84EE221 pKa = 4.22PEE223 pKa = 3.52HH224 pKa = 6.41RR225 pKa = 11.84AAVQRR230 pKa = 11.84NDD232 pKa = 2.96MNTLVNTVIRR242 pKa = 11.84EE243 pKa = 3.9VEE245 pKa = 3.8RR246 pKa = 11.84RR247 pKa = 11.84TKK249 pKa = 10.42RR250 pKa = 11.84EE251 pKa = 3.31RR252 pKa = 11.84LRR254 pKa = 11.84EE255 pKa = 3.98GRR257 pKa = 11.84RR258 pKa = 3.32
MM1 pKa = 7.41FFHH4 pKa = 6.36SWRR7 pKa = 11.84RR8 pKa = 11.84KK9 pKa = 8.67RR10 pKa = 11.84KK11 pKa = 8.75LKK13 pKa = 10.22RR14 pKa = 11.84FRR16 pKa = 11.84PIMTLHH22 pKa = 7.11LNRR25 pKa = 11.84TLHH28 pKa = 6.71HH29 pKa = 6.42EE30 pKa = 4.2FHH32 pKa = 6.56PTFLKK37 pKa = 10.84AKK39 pKa = 8.0TVLRR43 pKa = 11.84EE44 pKa = 3.85VRR46 pKa = 11.84SSVIRR51 pKa = 11.84KK52 pKa = 9.06EE53 pKa = 3.92EE54 pKa = 4.03HH55 pKa = 4.86THH57 pKa = 6.6HH58 pKa = 6.43IHH60 pKa = 7.73HH61 pKa = 6.71MMTLHH66 pKa = 5.69VHH68 pKa = 5.11QAQTFLMLNHH78 pKa = 6.12QNDD81 pKa = 3.69EE82 pKa = 4.08RR83 pKa = 11.84SLIIQSKK90 pKa = 9.78KK91 pKa = 10.05VINRR95 pKa = 11.84KK96 pKa = 8.59YY97 pKa = 9.95PSRR100 pKa = 11.84ILQLHH105 pKa = 6.41LAKK108 pKa = 10.53QSIEE112 pKa = 3.89RR113 pKa = 11.84LAEE116 pKa = 3.77QRR118 pKa = 11.84LLEE121 pKa = 4.3EE122 pKa = 3.72KK123 pKa = 10.25SYY125 pKa = 11.54YY126 pKa = 10.01KK127 pKa = 9.88YY128 pKa = 10.68HH129 pKa = 6.49PAKK132 pKa = 10.44AKK134 pKa = 9.84EE135 pKa = 3.81LAVSYY140 pKa = 9.22LHH142 pKa = 6.56NRR144 pKa = 11.84EE145 pKa = 4.17IKK147 pKa = 10.27NCLNTHH153 pKa = 5.9MNRR156 pKa = 11.84LSQYY160 pKa = 10.07SYY162 pKa = 10.89AGSLFRR168 pKa = 11.84SSLFTTLSSQSWKK181 pKa = 10.11QVLIPVIYY189 pKa = 8.79QQSRR193 pKa = 11.84ALIHH197 pKa = 6.43RR198 pKa = 11.84EE199 pKa = 3.63KK200 pKa = 10.52IQLQRR205 pKa = 11.84EE206 pKa = 4.52VISEE210 pKa = 4.02KK211 pKa = 10.83SRR213 pKa = 11.84DD214 pKa = 3.59EE215 pKa = 3.79MHH217 pKa = 6.56VEE219 pKa = 4.06RR220 pKa = 11.84EE221 pKa = 4.22PEE223 pKa = 3.52HH224 pKa = 6.41RR225 pKa = 11.84AAVQRR230 pKa = 11.84NDD232 pKa = 2.96MNTLVNTVIRR242 pKa = 11.84EE243 pKa = 3.9VEE245 pKa = 3.8RR246 pKa = 11.84RR247 pKa = 11.84TKK249 pKa = 10.42RR250 pKa = 11.84EE251 pKa = 3.31RR252 pKa = 11.84LRR254 pKa = 11.84EE255 pKa = 3.98GRR257 pKa = 11.84RR258 pKa = 3.32
Molecular weight: 31.35 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1343130 |
29 |
2670 |
288.7 |
32.69 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.181 ± 0.037 | 1.601 ± 0.014 |
5.703 ± 0.032 | 7.141 ± 0.041 |
4.197 ± 0.029 | 6.175 ± 0.043 |
2.278 ± 0.019 | 7.611 ± 0.034 |
6.825 ± 0.037 | 9.692 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.324 ± 0.019 | 4.227 ± 0.023 |
3.019 ± 0.019 | 3.958 ± 0.029 |
4.497 ± 0.028 | 5.796 ± 0.025 |
5.174 ± 0.03 | 6.427 ± 0.032 |
0.882 ± 0.011 | 4.292 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
