
Odonata-associated circular virus-8
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 7.71
Get precalculated fractions of proteins
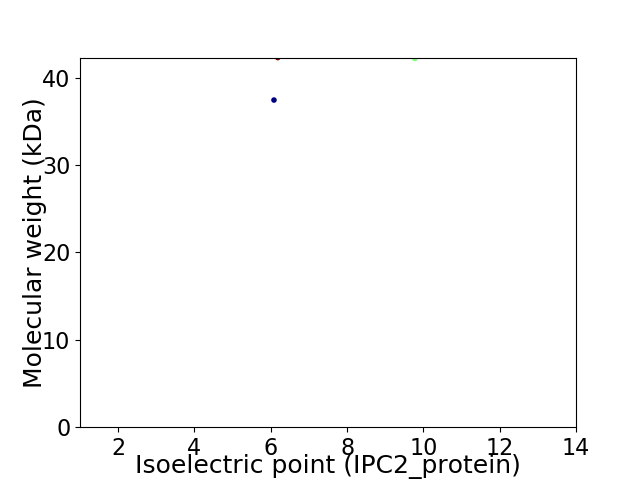
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
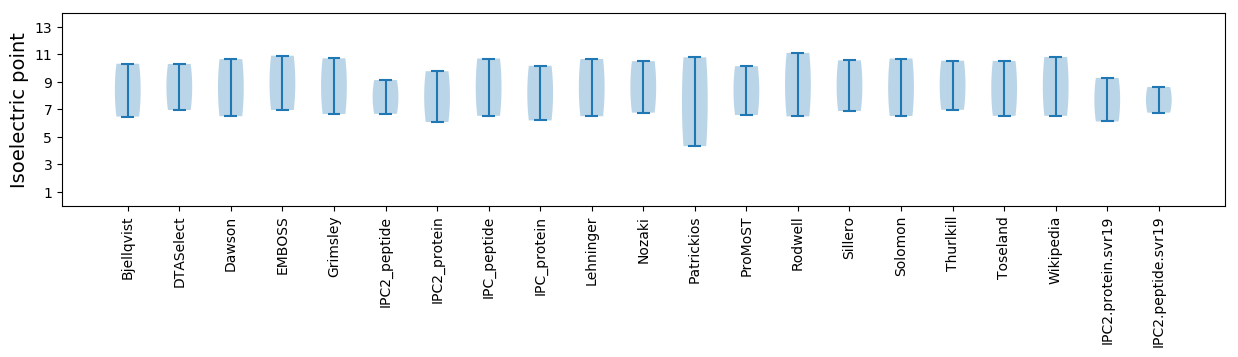
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B4UGS9|A0A0B4UGS9_9VIRU Replication-associated protein OS=Odonata-associated circular virus-8 OX=1592128 PE=3 SV=1
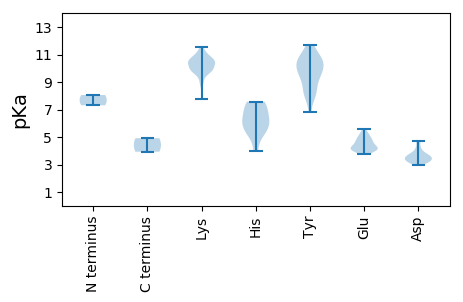
MM1 pKa = 8.06IEE3 pKa = 4.13DD4 pKa = 3.58VEE6 pKa = 4.85KK7 pKa = 11.06FFDD10 pKa = 3.69KK11 pKa = 10.52QDD13 pKa = 3.51KK14 pKa = 10.49KK15 pKa = 11.2KK16 pKa = 10.46EE17 pKa = 4.17GYY19 pKa = 9.61RR20 pKa = 11.84ATGKK24 pKa = 10.51AFFLTYY30 pKa = 9.05PKK32 pKa = 10.56CDD34 pKa = 3.25IKK36 pKa = 10.89RR37 pKa = 11.84DD38 pKa = 3.44EE39 pKa = 5.15AYY41 pKa = 10.07TMLAKK46 pKa = 9.96QWNPSNLIVAQEE58 pKa = 4.01DD59 pKa = 3.96HH60 pKa = 7.39KK61 pKa = 11.52DD62 pKa = 3.8GTPHH66 pKa = 6.29LHH68 pKa = 7.57VYY70 pKa = 8.13MQLPTKK76 pKa = 10.64KK77 pKa = 10.16NFKK80 pKa = 9.26NPRR83 pKa = 11.84WADD86 pKa = 3.12LGKK89 pKa = 10.22YY90 pKa = 9.21HH91 pKa = 7.49GFYY94 pKa = 8.15TTVRR98 pKa = 11.84DD99 pKa = 3.83RR100 pKa = 11.84QAVMAYY106 pKa = 9.71VMKK109 pKa = 10.81DD110 pKa = 3.08GNFKK114 pKa = 10.58MNEE117 pKa = 4.03NLNFTTWHH125 pKa = 6.25NFRR128 pKa = 11.84KK129 pKa = 10.11EE130 pKa = 3.96KK131 pKa = 10.87GDD133 pKa = 3.27FDD135 pKa = 3.3AWIIHH140 pKa = 5.77NEE142 pKa = 4.08SKK144 pKa = 10.58KK145 pKa = 10.85LKK147 pKa = 10.29SPYY150 pKa = 9.51PFKK153 pKa = 11.11LPDD156 pKa = 3.7GTDD159 pKa = 3.25VLEE162 pKa = 4.69PKK164 pKa = 10.56LGEE167 pKa = 4.17KK168 pKa = 9.81KK169 pKa = 9.83CNHH172 pKa = 6.61MIIGIPDD179 pKa = 3.39TGKK182 pKa = 10.46SFWYY186 pKa = 8.32NTTFEE191 pKa = 4.35GKK193 pKa = 9.77RR194 pKa = 11.84VFSRR198 pKa = 11.84VDD200 pKa = 2.96GKK202 pKa = 11.33YY203 pKa = 10.5PFDD206 pKa = 4.72DD207 pKa = 4.15YY208 pKa = 11.58EE209 pKa = 4.39GQEE212 pKa = 3.98VVIYY216 pKa = 10.41NDD218 pKa = 3.72PQTVTLMEE226 pKa = 5.61LINVSDD232 pKa = 4.65CYY234 pKa = 10.79LVKK237 pKa = 10.29TPVFGDD243 pKa = 3.06TRR245 pKa = 11.84YY246 pKa = 8.06TKK248 pKa = 10.26KK249 pKa = 9.6YY250 pKa = 8.63WKK252 pKa = 8.77MKK254 pKa = 7.8QRR256 pKa = 11.84RR257 pKa = 11.84TIIFICNHH265 pKa = 4.01GRR267 pKa = 11.84IPNFVEE273 pKa = 4.99DD274 pKa = 3.5PAFRR278 pKa = 11.84SRR280 pKa = 11.84FNIWDD285 pKa = 3.54MGDD288 pKa = 4.23DD289 pKa = 3.72EE290 pKa = 5.25PEE292 pKa = 4.41PEE294 pKa = 5.26EE295 pKa = 4.2MTDD298 pKa = 3.45DD299 pKa = 4.14PNRR302 pKa = 11.84TEE304 pKa = 4.08MDD306 pKa = 3.22TDD308 pKa = 4.59FDD310 pKa = 3.79ATDD313 pKa = 3.45YY314 pKa = 11.66ARR316 pKa = 4.91
MM1 pKa = 8.06IEE3 pKa = 4.13DD4 pKa = 3.58VEE6 pKa = 4.85KK7 pKa = 11.06FFDD10 pKa = 3.69KK11 pKa = 10.52QDD13 pKa = 3.51KK14 pKa = 10.49KK15 pKa = 11.2KK16 pKa = 10.46EE17 pKa = 4.17GYY19 pKa = 9.61RR20 pKa = 11.84ATGKK24 pKa = 10.51AFFLTYY30 pKa = 9.05PKK32 pKa = 10.56CDD34 pKa = 3.25IKK36 pKa = 10.89RR37 pKa = 11.84DD38 pKa = 3.44EE39 pKa = 5.15AYY41 pKa = 10.07TMLAKK46 pKa = 9.96QWNPSNLIVAQEE58 pKa = 4.01DD59 pKa = 3.96HH60 pKa = 7.39KK61 pKa = 11.52DD62 pKa = 3.8GTPHH66 pKa = 6.29LHH68 pKa = 7.57VYY70 pKa = 8.13MQLPTKK76 pKa = 10.64KK77 pKa = 10.16NFKK80 pKa = 9.26NPRR83 pKa = 11.84WADD86 pKa = 3.12LGKK89 pKa = 10.22YY90 pKa = 9.21HH91 pKa = 7.49GFYY94 pKa = 8.15TTVRR98 pKa = 11.84DD99 pKa = 3.83RR100 pKa = 11.84QAVMAYY106 pKa = 9.71VMKK109 pKa = 10.81DD110 pKa = 3.08GNFKK114 pKa = 10.58MNEE117 pKa = 4.03NLNFTTWHH125 pKa = 6.25NFRR128 pKa = 11.84KK129 pKa = 10.11EE130 pKa = 3.96KK131 pKa = 10.87GDD133 pKa = 3.27FDD135 pKa = 3.3AWIIHH140 pKa = 5.77NEE142 pKa = 4.08SKK144 pKa = 10.58KK145 pKa = 10.85LKK147 pKa = 10.29SPYY150 pKa = 9.51PFKK153 pKa = 11.11LPDD156 pKa = 3.7GTDD159 pKa = 3.25VLEE162 pKa = 4.69PKK164 pKa = 10.56LGEE167 pKa = 4.17KK168 pKa = 9.81KK169 pKa = 9.83CNHH172 pKa = 6.61MIIGIPDD179 pKa = 3.39TGKK182 pKa = 10.46SFWYY186 pKa = 8.32NTTFEE191 pKa = 4.35GKK193 pKa = 9.77RR194 pKa = 11.84VFSRR198 pKa = 11.84VDD200 pKa = 2.96GKK202 pKa = 11.33YY203 pKa = 10.5PFDD206 pKa = 4.72DD207 pKa = 4.15YY208 pKa = 11.58EE209 pKa = 4.39GQEE212 pKa = 3.98VVIYY216 pKa = 10.41NDD218 pKa = 3.72PQTVTLMEE226 pKa = 5.61LINVSDD232 pKa = 4.65CYY234 pKa = 10.79LVKK237 pKa = 10.29TPVFGDD243 pKa = 3.06TRR245 pKa = 11.84YY246 pKa = 8.06TKK248 pKa = 10.26KK249 pKa = 9.6YY250 pKa = 8.63WKK252 pKa = 8.77MKK254 pKa = 7.8QRR256 pKa = 11.84RR257 pKa = 11.84TIIFICNHH265 pKa = 4.01GRR267 pKa = 11.84IPNFVEE273 pKa = 4.99DD274 pKa = 3.5PAFRR278 pKa = 11.84SRR280 pKa = 11.84FNIWDD285 pKa = 3.54MGDD288 pKa = 4.23DD289 pKa = 3.72EE290 pKa = 5.25PEE292 pKa = 4.41PEE294 pKa = 5.26EE295 pKa = 4.2MTDD298 pKa = 3.45DD299 pKa = 4.14PNRR302 pKa = 11.84TEE304 pKa = 4.08MDD306 pKa = 3.22TDD308 pKa = 4.59FDD310 pKa = 3.79ATDD313 pKa = 3.45YY314 pKa = 11.66ARR316 pKa = 4.91
Molecular weight: 37.44 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B4UGS9|A0A0B4UGS9_9VIRU Replication-associated protein OS=Odonata-associated circular virus-8 OX=1592128 PE=3 SV=1
MM1 pKa = 7.31SKK3 pKa = 10.62AAGHH7 pKa = 7.22LATTLAGLDD16 pKa = 3.36QIIRR20 pKa = 11.84SGKK23 pKa = 9.07SMYY26 pKa = 10.56NRR28 pKa = 11.84FSNWRR33 pKa = 11.84NPRR36 pKa = 11.84QVTPTRR42 pKa = 11.84SFPKK46 pKa = 9.41FSTRR50 pKa = 11.84MRR52 pKa = 11.84TISRR56 pKa = 11.84RR57 pKa = 11.84SFTKK61 pKa = 10.13SRR63 pKa = 11.84SSNGTRR69 pKa = 11.84KK70 pKa = 9.71RR71 pKa = 11.84SYY73 pKa = 9.16NGKK76 pKa = 9.66RR77 pKa = 11.84KK78 pKa = 9.88RR79 pKa = 11.84NSTKK83 pKa = 8.85RR84 pKa = 11.84TVWRR88 pKa = 11.84MKK90 pKa = 10.58KK91 pKa = 9.8KK92 pKa = 10.23LKK94 pKa = 10.45QIMVPAKK101 pKa = 10.41SLWLDD106 pKa = 3.47ALQGRR111 pKa = 11.84VTTGQGTSTWVSLGNLLTQNAVLTGFTGSVEE142 pKa = 4.12SGTYY146 pKa = 10.62LMSGLANNDD155 pKa = 3.23GTSGNILGRR164 pKa = 11.84AKK166 pKa = 10.61FIVHH170 pKa = 5.32SWEE173 pKa = 4.51QIFRR177 pKa = 11.84WTNQSNCPLTLEE189 pKa = 4.92CYY191 pKa = 7.95TLKK194 pKa = 10.56CVQNVPVDD202 pKa = 3.58GLINSGLEE210 pKa = 3.79VTNTIKK216 pKa = 11.09DD217 pKa = 3.69FMTACYY223 pKa = 9.9NKK225 pKa = 9.88QYY227 pKa = 10.97VGGSTIPSISVDD239 pKa = 3.15HH240 pKa = 6.87PAFKK244 pKa = 10.83LSDD247 pKa = 3.6LARR250 pKa = 11.84FGAYY254 pKa = 9.7FKK256 pKa = 10.13TIRR259 pKa = 11.84HH260 pKa = 5.15KK261 pKa = 9.79TVVVQPGEE269 pKa = 4.23SMSMRR274 pKa = 11.84KK275 pKa = 9.78YY276 pKa = 10.33KK277 pKa = 10.1IRR279 pKa = 11.84PRR281 pKa = 11.84TYY283 pKa = 10.95AEE285 pKa = 4.7AISYY289 pKa = 10.01DD290 pKa = 3.39AVTAVLKK297 pKa = 10.04VHH299 pKa = 5.75AQKK302 pKa = 9.85GTLHH306 pKa = 5.56YY307 pKa = 9.88LWRR310 pKa = 11.84ITGTPQSAAEE320 pKa = 4.41GKK322 pKa = 9.96DD323 pKa = 3.14ATLCDD328 pKa = 3.36VKK330 pKa = 10.53MAFVSSIHH338 pKa = 6.25ARR340 pKa = 11.84TSGYY344 pKa = 10.66SSNNYY349 pKa = 6.8QTFPSTTGLPVGLVIEE365 pKa = 4.75NIYY368 pKa = 10.5PGTSEE373 pKa = 4.75AKK375 pKa = 9.79VAAPVPP381 pKa = 3.93
MM1 pKa = 7.31SKK3 pKa = 10.62AAGHH7 pKa = 7.22LATTLAGLDD16 pKa = 3.36QIIRR20 pKa = 11.84SGKK23 pKa = 9.07SMYY26 pKa = 10.56NRR28 pKa = 11.84FSNWRR33 pKa = 11.84NPRR36 pKa = 11.84QVTPTRR42 pKa = 11.84SFPKK46 pKa = 9.41FSTRR50 pKa = 11.84MRR52 pKa = 11.84TISRR56 pKa = 11.84RR57 pKa = 11.84SFTKK61 pKa = 10.13SRR63 pKa = 11.84SSNGTRR69 pKa = 11.84KK70 pKa = 9.71RR71 pKa = 11.84SYY73 pKa = 9.16NGKK76 pKa = 9.66RR77 pKa = 11.84KK78 pKa = 9.88RR79 pKa = 11.84NSTKK83 pKa = 8.85RR84 pKa = 11.84TVWRR88 pKa = 11.84MKK90 pKa = 10.58KK91 pKa = 9.8KK92 pKa = 10.23LKK94 pKa = 10.45QIMVPAKK101 pKa = 10.41SLWLDD106 pKa = 3.47ALQGRR111 pKa = 11.84VTTGQGTSTWVSLGNLLTQNAVLTGFTGSVEE142 pKa = 4.12SGTYY146 pKa = 10.62LMSGLANNDD155 pKa = 3.23GTSGNILGRR164 pKa = 11.84AKK166 pKa = 10.61FIVHH170 pKa = 5.32SWEE173 pKa = 4.51QIFRR177 pKa = 11.84WTNQSNCPLTLEE189 pKa = 4.92CYY191 pKa = 7.95TLKK194 pKa = 10.56CVQNVPVDD202 pKa = 3.58GLINSGLEE210 pKa = 3.79VTNTIKK216 pKa = 11.09DD217 pKa = 3.69FMTACYY223 pKa = 9.9NKK225 pKa = 9.88QYY227 pKa = 10.97VGGSTIPSISVDD239 pKa = 3.15HH240 pKa = 6.87PAFKK244 pKa = 10.83LSDD247 pKa = 3.6LARR250 pKa = 11.84FGAYY254 pKa = 9.7FKK256 pKa = 10.13TIRR259 pKa = 11.84HH260 pKa = 5.15KK261 pKa = 9.79TVVVQPGEE269 pKa = 4.23SMSMRR274 pKa = 11.84KK275 pKa = 9.78YY276 pKa = 10.33KK277 pKa = 10.1IRR279 pKa = 11.84PRR281 pKa = 11.84TYY283 pKa = 10.95AEE285 pKa = 4.7AISYY289 pKa = 10.01DD290 pKa = 3.39AVTAVLKK297 pKa = 10.04VHH299 pKa = 5.75AQKK302 pKa = 9.85GTLHH306 pKa = 5.56YY307 pKa = 9.88LWRR310 pKa = 11.84ITGTPQSAAEE320 pKa = 4.41GKK322 pKa = 9.96DD323 pKa = 3.14ATLCDD328 pKa = 3.36VKK330 pKa = 10.53MAFVSSIHH338 pKa = 6.25ARR340 pKa = 11.84TSGYY344 pKa = 10.66SSNNYY349 pKa = 6.8QTFPSTTGLPVGLVIEE365 pKa = 4.75NIYY368 pKa = 10.5PGTSEE373 pKa = 4.75AKK375 pKa = 9.79VAAPVPP381 pKa = 3.93
Molecular weight: 42.2 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
697 |
316 |
381 |
348.5 |
39.82 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.452 ± 1.084 | 1.291 ± 0.017 |
5.882 ± 2.573 | 4.161 ± 1.421 |
4.878 ± 1.158 | 6.6 ± 0.799 |
2.152 ± 0.249 | 4.735 ± 0.008 |
8.608 ± 1.202 | 5.882 ± 0.951 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.156 ± 0.42 | 5.452 ± 0.16 |
4.878 ± 0.536 | 3.156 ± 0.409 |
6.026 ± 0.631 | 6.456 ± 2.778 |
8.895 ± 1.059 | 6.026 ± 0.631 |
2.009 ± 0.135 | 4.304 ± 0.497 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
