
Heliorestis convoluta
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Heliobacteriaceae; Heliorestis
Average proteome isoelectric point is 6.5
Get precalculated fractions of proteins
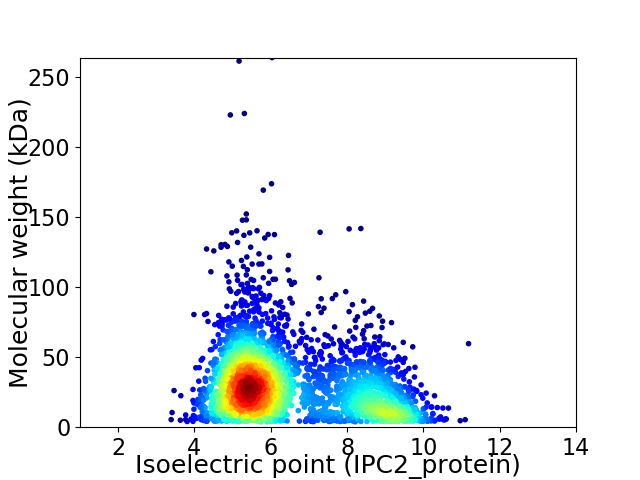
Virtual 2D-PAGE plot for 3259 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5Q2MW37|A0A5Q2MW37_9FIRM Copper amine oxidase N-terminal domain-containing protein OS=Heliorestis convoluta OX=356322 GN=FTV88_0288 PE=4 SV=1
MM1 pKa = 7.87PSLKK5 pKa = 10.29EE6 pKa = 3.75RR7 pKa = 11.84ISYY10 pKa = 9.36IKK12 pKa = 10.77GMAEE16 pKa = 3.83GMKK19 pKa = 9.92IEE21 pKa = 4.43ADD23 pKa = 3.42PRR25 pKa = 11.84EE26 pKa = 4.05GRR28 pKa = 11.84LLSEE32 pKa = 5.2MITILDD38 pKa = 3.57TMAEE42 pKa = 4.16EE43 pKa = 4.14IEE45 pKa = 4.27EE46 pKa = 4.06VRR48 pKa = 11.84RR49 pKa = 11.84NQASLEE55 pKa = 4.01EE56 pKa = 4.24MVEE59 pKa = 4.48SIDD62 pKa = 3.8EE63 pKa = 4.26DD64 pKa = 4.65LVDD67 pKa = 5.54LEE69 pKa = 4.27NDD71 pKa = 3.72FYY73 pKa = 11.71DD74 pKa = 5.29LEE76 pKa = 5.18EE77 pKa = 6.36DD78 pKa = 5.03DD79 pKa = 6.72FDD81 pKa = 7.01DD82 pKa = 5.84EE83 pKa = 6.51DD84 pKa = 6.34FDD86 pKa = 7.55DD87 pKa = 6.47DD88 pKa = 5.46DD89 pKa = 7.21DD90 pKa = 7.32DD91 pKa = 7.33GILCDD96 pKa = 5.91CDD98 pKa = 6.09DD99 pKa = 5.8DD100 pKa = 7.08DD101 pKa = 7.43DD102 pKa = 5.51DD103 pKa = 5.8VIEE106 pKa = 5.21FEE108 pKa = 5.64CPTCNEE114 pKa = 4.27TVCFEE119 pKa = 4.64EE120 pKa = 5.23EE121 pKa = 4.51DD122 pKa = 4.1LDD124 pKa = 4.46DD125 pKa = 5.99DD126 pKa = 4.09EE127 pKa = 5.03VLEE130 pKa = 4.77VVCPTCDD137 pKa = 2.94HH138 pKa = 6.14TVYY141 pKa = 10.86VQDD144 pKa = 5.69GEE146 pKa = 4.43DD147 pKa = 4.17LDD149 pKa = 4.17MTACDD154 pKa = 5.76DD155 pKa = 4.49DD156 pKa = 7.13DD157 pKa = 4.97EE158 pKa = 6.21DD159 pKa = 5.07SPCFRR164 pKa = 11.84NNEE167 pKa = 3.88EE168 pKa = 4.03ATRR171 pKa = 11.84IPQSRR176 pKa = 11.84PPRR179 pKa = 11.84EE180 pKa = 3.95TYY182 pKa = 9.7RR183 pKa = 11.84ARR185 pKa = 11.84PYY187 pKa = 10.32HH188 pKa = 4.95YY189 pKa = 9.81HH190 pKa = 5.62HH191 pKa = 7.28HH192 pKa = 5.9NN193 pKa = 3.22
MM1 pKa = 7.87PSLKK5 pKa = 10.29EE6 pKa = 3.75RR7 pKa = 11.84ISYY10 pKa = 9.36IKK12 pKa = 10.77GMAEE16 pKa = 3.83GMKK19 pKa = 9.92IEE21 pKa = 4.43ADD23 pKa = 3.42PRR25 pKa = 11.84EE26 pKa = 4.05GRR28 pKa = 11.84LLSEE32 pKa = 5.2MITILDD38 pKa = 3.57TMAEE42 pKa = 4.16EE43 pKa = 4.14IEE45 pKa = 4.27EE46 pKa = 4.06VRR48 pKa = 11.84RR49 pKa = 11.84NQASLEE55 pKa = 4.01EE56 pKa = 4.24MVEE59 pKa = 4.48SIDD62 pKa = 3.8EE63 pKa = 4.26DD64 pKa = 4.65LVDD67 pKa = 5.54LEE69 pKa = 4.27NDD71 pKa = 3.72FYY73 pKa = 11.71DD74 pKa = 5.29LEE76 pKa = 5.18EE77 pKa = 6.36DD78 pKa = 5.03DD79 pKa = 6.72FDD81 pKa = 7.01DD82 pKa = 5.84EE83 pKa = 6.51DD84 pKa = 6.34FDD86 pKa = 7.55DD87 pKa = 6.47DD88 pKa = 5.46DD89 pKa = 7.21DD90 pKa = 7.32DD91 pKa = 7.33GILCDD96 pKa = 5.91CDD98 pKa = 6.09DD99 pKa = 5.8DD100 pKa = 7.08DD101 pKa = 7.43DD102 pKa = 5.51DD103 pKa = 5.8VIEE106 pKa = 5.21FEE108 pKa = 5.64CPTCNEE114 pKa = 4.27TVCFEE119 pKa = 4.64EE120 pKa = 5.23EE121 pKa = 4.51DD122 pKa = 4.1LDD124 pKa = 4.46DD125 pKa = 5.99DD126 pKa = 4.09EE127 pKa = 5.03VLEE130 pKa = 4.77VVCPTCDD137 pKa = 2.94HH138 pKa = 6.14TVYY141 pKa = 10.86VQDD144 pKa = 5.69GEE146 pKa = 4.43DD147 pKa = 4.17LDD149 pKa = 4.17MTACDD154 pKa = 5.76DD155 pKa = 4.49DD156 pKa = 7.13DD157 pKa = 4.97EE158 pKa = 6.21DD159 pKa = 5.07SPCFRR164 pKa = 11.84NNEE167 pKa = 3.88EE168 pKa = 4.03ATRR171 pKa = 11.84IPQSRR176 pKa = 11.84PPRR179 pKa = 11.84EE180 pKa = 3.95TYY182 pKa = 9.7RR183 pKa = 11.84ARR185 pKa = 11.84PYY187 pKa = 10.32HH188 pKa = 4.95YY189 pKa = 9.81HH190 pKa = 5.62HH191 pKa = 7.28HH192 pKa = 5.9NN193 pKa = 3.22
Molecular weight: 22.49 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5Q2N406|A0A5Q2N406_9FIRM Nickel pincer cofactor biosynthesis protein LarC OS=Heliorestis convoluta OX=356322 GN=FTV88_0843 PE=4 SV=1
MM1 pKa = 7.32MAPLHH6 pKa = 6.65HH7 pKa = 6.71RR8 pKa = 11.84TQQRR12 pKa = 11.84IINQNPRR19 pKa = 11.84LHH21 pKa = 6.62ILNPLRR27 pKa = 11.84PMKK30 pKa = 10.42KK31 pKa = 8.94IASLHH36 pKa = 5.96SMHH39 pKa = 7.17LNPIIPLMRR48 pKa = 11.84QVRR51 pKa = 11.84ILKK54 pKa = 8.17TRR56 pKa = 11.84NHH58 pKa = 6.6QIHH61 pKa = 7.1PMPLIQKK68 pKa = 8.53PLHH71 pKa = 6.41NPLCMKK77 pKa = 10.07RR78 pKa = 11.84PRR80 pKa = 11.84RR81 pKa = 11.84ILGIIKK87 pKa = 8.56ITNNNNTHH95 pKa = 4.69QTTTSFSRR103 pKa = 11.84KK104 pKa = 6.82ITHH107 pKa = 6.93KK108 pKa = 10.29GTVLPPDD115 pKa = 4.04PPRR118 pKa = 11.84INHH121 pKa = 6.44NLLLQNLPIHH131 pKa = 6.45KK132 pKa = 9.41SKK134 pKa = 11.4SPMIYY139 pKa = 9.62QKK141 pKa = 10.35HH142 pKa = 5.99HH143 pKa = 6.84NISSLNRR150 pKa = 11.84LPHH153 pKa = 5.41IRR155 pKa = 11.84KK156 pKa = 7.28MHH158 pKa = 6.37TIITSQRR165 pKa = 11.84SRR167 pKa = 11.84QRR169 pKa = 11.84RR170 pKa = 11.84NMRR173 pKa = 11.84LHH175 pKa = 6.75HH176 pKa = 6.85IKK178 pKa = 10.16LAHH181 pKa = 6.54LVRR184 pKa = 11.84SQMTHH189 pKa = 5.27NLQSRR194 pKa = 11.84TLPQIINISLKK205 pKa = 10.5RR206 pKa = 11.84KK207 pKa = 7.15PQQRR211 pKa = 11.84NHH213 pKa = 6.41RR214 pKa = 11.84SAKK217 pKa = 7.67TLRR220 pKa = 11.84LLLNLTQHH228 pKa = 6.3MKK230 pKa = 10.32RR231 pKa = 11.84LPIIHH236 pKa = 6.65IPSRR240 pKa = 11.84PNQPRR245 pKa = 11.84LLRR248 pKa = 11.84RR249 pKa = 11.84RR250 pKa = 11.84IHH252 pKa = 5.8NKK254 pKa = 9.15PRR256 pKa = 11.84IHH258 pKa = 7.04CNTMTTHH265 pKa = 6.18PRR267 pKa = 11.84SRR269 pKa = 11.84LQNTHH274 pKa = 5.55PRR276 pKa = 11.84MAIGQINQFPHH287 pKa = 7.14IHH289 pKa = 6.02IQTLANQRR297 pKa = 11.84KK298 pKa = 9.56LIGKK302 pKa = 9.01SNIHH306 pKa = 4.4ITKK309 pKa = 10.47RR310 pKa = 11.84ILRR313 pKa = 11.84QLRR316 pKa = 11.84QLRR319 pKa = 11.84RR320 pKa = 11.84PSRR323 pKa = 11.84RR324 pKa = 11.84QQNLPLHH331 pKa = 6.12KK332 pKa = 10.32RR333 pKa = 11.84LIKK336 pKa = 9.54KK337 pKa = 7.61TSPLRR342 pKa = 11.84RR343 pKa = 11.84SQINAAHH350 pKa = 6.2NPVVINQLPQNLPRR364 pKa = 11.84QHH366 pKa = 7.5PLRR369 pKa = 11.84TMRR372 pKa = 11.84QIHH375 pKa = 6.7LPRR378 pKa = 11.84HH379 pKa = 4.49LQIRR383 pKa = 11.84PQLPQPLSNRR393 pKa = 11.84LCRR396 pKa = 11.84SHH398 pKa = 6.49RR399 pKa = 11.84TGRR402 pKa = 11.84LQNNQISRR410 pKa = 11.84LYY412 pKa = 9.97IGRR415 pKa = 11.84NSTRR419 pKa = 11.84RR420 pKa = 11.84RR421 pKa = 11.84LNIRR425 pKa = 11.84KK426 pKa = 8.65IRR428 pKa = 11.84HH429 pKa = 4.93MPLLKK434 pKa = 10.05RR435 pKa = 11.84RR436 pKa = 11.84RR437 pKa = 11.84HH438 pKa = 5.12RR439 pKa = 11.84QNKK442 pKa = 8.93NISLFRR448 pKa = 11.84IHH450 pKa = 7.65RR451 pKa = 11.84SLQIPTMDD459 pKa = 3.81SRR461 pKa = 11.84LHH463 pKa = 5.26QNIQIRR469 pKa = 11.84LINMDD474 pKa = 3.72LTTINRR480 pKa = 11.84LHH482 pKa = 6.84SLRR485 pKa = 11.84IHH487 pKa = 6.54INTMNTKK494 pKa = 7.51TLRR497 pKa = 11.84RR498 pKa = 3.8
MM1 pKa = 7.32MAPLHH6 pKa = 6.65HH7 pKa = 6.71RR8 pKa = 11.84TQQRR12 pKa = 11.84IINQNPRR19 pKa = 11.84LHH21 pKa = 6.62ILNPLRR27 pKa = 11.84PMKK30 pKa = 10.42KK31 pKa = 8.94IASLHH36 pKa = 5.96SMHH39 pKa = 7.17LNPIIPLMRR48 pKa = 11.84QVRR51 pKa = 11.84ILKK54 pKa = 8.17TRR56 pKa = 11.84NHH58 pKa = 6.6QIHH61 pKa = 7.1PMPLIQKK68 pKa = 8.53PLHH71 pKa = 6.41NPLCMKK77 pKa = 10.07RR78 pKa = 11.84PRR80 pKa = 11.84RR81 pKa = 11.84ILGIIKK87 pKa = 8.56ITNNNNTHH95 pKa = 4.69QTTTSFSRR103 pKa = 11.84KK104 pKa = 6.82ITHH107 pKa = 6.93KK108 pKa = 10.29GTVLPPDD115 pKa = 4.04PPRR118 pKa = 11.84INHH121 pKa = 6.44NLLLQNLPIHH131 pKa = 6.45KK132 pKa = 9.41SKK134 pKa = 11.4SPMIYY139 pKa = 9.62QKK141 pKa = 10.35HH142 pKa = 5.99HH143 pKa = 6.84NISSLNRR150 pKa = 11.84LPHH153 pKa = 5.41IRR155 pKa = 11.84KK156 pKa = 7.28MHH158 pKa = 6.37TIITSQRR165 pKa = 11.84SRR167 pKa = 11.84QRR169 pKa = 11.84RR170 pKa = 11.84NMRR173 pKa = 11.84LHH175 pKa = 6.75HH176 pKa = 6.85IKK178 pKa = 10.16LAHH181 pKa = 6.54LVRR184 pKa = 11.84SQMTHH189 pKa = 5.27NLQSRR194 pKa = 11.84TLPQIINISLKK205 pKa = 10.5RR206 pKa = 11.84KK207 pKa = 7.15PQQRR211 pKa = 11.84NHH213 pKa = 6.41RR214 pKa = 11.84SAKK217 pKa = 7.67TLRR220 pKa = 11.84LLLNLTQHH228 pKa = 6.3MKK230 pKa = 10.32RR231 pKa = 11.84LPIIHH236 pKa = 6.65IPSRR240 pKa = 11.84PNQPRR245 pKa = 11.84LLRR248 pKa = 11.84RR249 pKa = 11.84RR250 pKa = 11.84IHH252 pKa = 5.8NKK254 pKa = 9.15PRR256 pKa = 11.84IHH258 pKa = 7.04CNTMTTHH265 pKa = 6.18PRR267 pKa = 11.84SRR269 pKa = 11.84LQNTHH274 pKa = 5.55PRR276 pKa = 11.84MAIGQINQFPHH287 pKa = 7.14IHH289 pKa = 6.02IQTLANQRR297 pKa = 11.84KK298 pKa = 9.56LIGKK302 pKa = 9.01SNIHH306 pKa = 4.4ITKK309 pKa = 10.47RR310 pKa = 11.84ILRR313 pKa = 11.84QLRR316 pKa = 11.84QLRR319 pKa = 11.84RR320 pKa = 11.84PSRR323 pKa = 11.84RR324 pKa = 11.84QQNLPLHH331 pKa = 6.12KK332 pKa = 10.32RR333 pKa = 11.84LIKK336 pKa = 9.54KK337 pKa = 7.61TSPLRR342 pKa = 11.84RR343 pKa = 11.84SQINAAHH350 pKa = 6.2NPVVINQLPQNLPRR364 pKa = 11.84QHH366 pKa = 7.5PLRR369 pKa = 11.84TMRR372 pKa = 11.84QIHH375 pKa = 6.7LPRR378 pKa = 11.84HH379 pKa = 4.49LQIRR383 pKa = 11.84PQLPQPLSNRR393 pKa = 11.84LCRR396 pKa = 11.84SHH398 pKa = 6.49RR399 pKa = 11.84TGRR402 pKa = 11.84LQNNQISRR410 pKa = 11.84LYY412 pKa = 9.97IGRR415 pKa = 11.84NSTRR419 pKa = 11.84RR420 pKa = 11.84RR421 pKa = 11.84LNIRR425 pKa = 11.84KK426 pKa = 8.65IRR428 pKa = 11.84HH429 pKa = 4.93MPLLKK434 pKa = 10.05RR435 pKa = 11.84RR436 pKa = 11.84RR437 pKa = 11.84HH438 pKa = 5.12RR439 pKa = 11.84QNKK442 pKa = 8.93NISLFRR448 pKa = 11.84IHH450 pKa = 7.65RR451 pKa = 11.84SLQIPTMDD459 pKa = 3.81SRR461 pKa = 11.84LHH463 pKa = 5.26QNIQIRR469 pKa = 11.84LINMDD474 pKa = 3.72LTTINRR480 pKa = 11.84LHH482 pKa = 6.84SLRR485 pKa = 11.84IHH487 pKa = 6.54INTMNTKK494 pKa = 7.51TLRR497 pKa = 11.84RR498 pKa = 3.8
Molecular weight: 59.65 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
928659 |
31 |
2326 |
285.0 |
31.96 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.907 ± 0.053 | 0.965 ± 0.022 |
4.871 ± 0.029 | 7.734 ± 0.053 |
3.882 ± 0.029 | 6.823 ± 0.048 |
2.078 ± 0.019 | 7.045 ± 0.034 |
6.118 ± 0.049 | 10.322 ± 0.058 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.603 ± 0.02 | 3.665 ± 0.039 |
4.029 ± 0.031 | 4.534 ± 0.049 |
5.342 ± 0.037 | 5.771 ± 0.032 |
5.211 ± 0.035 | 6.811 ± 0.039 |
1.074 ± 0.018 | 3.216 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
