
Mycobacterium phage Mdavu
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; Timquatrovirus; unclassified Timquatrovirus
Average proteome isoelectric point is 6.17
Get precalculated fractions of proteins
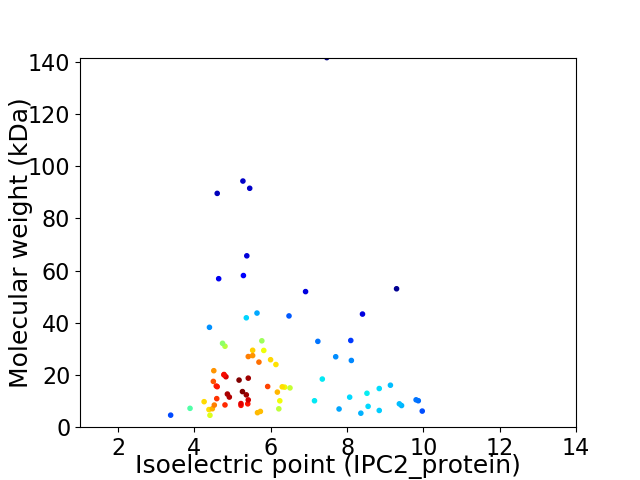
Virtual 2D-PAGE plot for 78 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
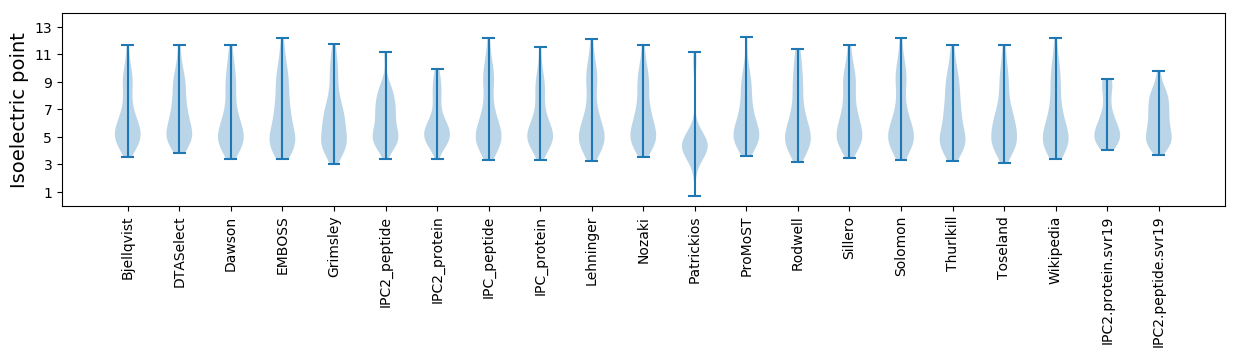
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A649VLU8|A0A649VLU8_9CAUD Uncharacterized protein OS=Mycobacterium phage Mdavu OX=2656595 GN=33 PE=4 SV=1
MM1 pKa = 8.09DD2 pKa = 5.43APTHH6 pKa = 5.1FTAVASTGDD15 pKa = 3.51EE16 pKa = 4.33PGWLLVHH23 pKa = 6.26VPEE26 pKa = 5.77IDD28 pKa = 3.29QYY30 pKa = 9.19TQARR34 pKa = 11.84TLDD37 pKa = 3.64EE38 pKa = 4.8VGPMAADD45 pKa = 6.26LIATWLNVPVEE56 pKa = 4.4SVDD59 pKa = 4.08VEE61 pKa = 4.63VKK63 pKa = 9.71YY64 pKa = 9.62TDD66 pKa = 3.01
MM1 pKa = 8.09DD2 pKa = 5.43APTHH6 pKa = 5.1FTAVASTGDD15 pKa = 3.51EE16 pKa = 4.33PGWLLVHH23 pKa = 6.26VPEE26 pKa = 5.77IDD28 pKa = 3.29QYY30 pKa = 9.19TQARR34 pKa = 11.84TLDD37 pKa = 3.64EE38 pKa = 4.8VGPMAADD45 pKa = 6.26LIATWLNVPVEE56 pKa = 4.4SVDD59 pKa = 4.08VEE61 pKa = 4.63VKK63 pKa = 9.71YY64 pKa = 9.62TDD66 pKa = 3.01
Molecular weight: 7.23 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A649VN02|A0A649VN02_9CAUD Uncharacterized protein OS=Mycobacterium phage Mdavu OX=2656595 GN=48 PE=4 SV=1
MM1 pKa = 7.0PCYY4 pKa = 10.12RR5 pKa = 11.84RR6 pKa = 11.84THH8 pKa = 5.41GFEE11 pKa = 4.33LRR13 pKa = 11.84ICVGTGARR21 pKa = 11.84GGPRR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84SQQRR31 pKa = 11.84VQSRR35 pKa = 11.84SEE37 pKa = 4.15CVADD41 pKa = 3.0IHH43 pKa = 6.48TPLVVLGIWTFFHH56 pKa = 5.62VVSRR60 pKa = 11.84AVNRR64 pKa = 11.84CFRR67 pKa = 11.84RR68 pKa = 11.84SVPIYY73 pKa = 10.0DD74 pKa = 3.87QISIPGSSTLKK85 pKa = 10.9SPGQRR90 pKa = 11.84HH91 pKa = 5.46FLWPGLFYY99 pKa = 10.75IHH101 pKa = 6.57TPSHH105 pKa = 6.19KK106 pKa = 9.78SLRR109 pKa = 11.84SATMASLRR117 pKa = 11.84TGSRR121 pKa = 11.84KK122 pKa = 10.39DD123 pKa = 3.1GTTYY127 pKa = 8.99TQVRR131 pKa = 11.84YY132 pKa = 9.85RR133 pKa = 11.84LNGKK137 pKa = 7.22QTSTSFDD144 pKa = 3.6DD145 pKa = 3.74PAQAVEE151 pKa = 4.86FKK153 pKa = 11.16RR154 pKa = 11.84MVEE157 pKa = 3.72QLGAAKK163 pKa = 9.9ALEE166 pKa = 4.61VIEE169 pKa = 4.39STEE172 pKa = 3.75ATAQHH177 pKa = 5.66YY178 pKa = 7.57TVGDD182 pKa = 3.64WLDD185 pKa = 3.79HH186 pKa = 5.61YY187 pKa = 10.59LSHH190 pKa = 6.37KK191 pKa = 9.54TGVEE195 pKa = 3.53KK196 pKa = 10.38STMYY200 pKa = 10.58DD201 pKa = 3.23YY202 pKa = 11.63RR203 pKa = 11.84KK204 pKa = 9.14MVEE207 pKa = 3.85KK208 pKa = 10.62DD209 pKa = 2.84IAPVLGPIPLAALTAEE225 pKa = 4.6DD226 pKa = 3.03VAMWVKK232 pKa = 10.63GLAARR237 pKa = 11.84GLKK240 pKa = 10.35GKK242 pKa = 8.65TIANKK247 pKa = 10.32HH248 pKa = 5.45GFLSSALKK256 pKa = 10.03VAASRR261 pKa = 11.84GLIKK265 pKa = 10.8SNPAVGGVGMVEE277 pKa = 4.11IPRR280 pKa = 11.84SEE282 pKa = 4.0RR283 pKa = 11.84EE284 pKa = 3.63EE285 pKa = 4.01MVFLTRR291 pKa = 11.84EE292 pKa = 3.65QYY294 pKa = 11.16ARR296 pKa = 11.84LRR298 pKa = 11.84DD299 pKa = 4.07CVTEE303 pKa = 3.8PWRR306 pKa = 11.84PLVEE310 pKa = 4.14FLVASGARR318 pKa = 11.84WGEE321 pKa = 4.04VTALRR326 pKa = 11.84PSDD329 pKa = 3.63VNRR332 pKa = 11.84DD333 pKa = 3.36EE334 pKa = 4.11GTVRR338 pKa = 11.84ISRR341 pKa = 11.84AWKK344 pKa = 8.46RR345 pKa = 11.84TYY347 pKa = 8.71EE348 pKa = 4.0TGGYY352 pKa = 9.78EE353 pKa = 3.6IGAPKK358 pKa = 9.2TDD360 pKa = 3.04KK361 pKa = 10.73SRR363 pKa = 11.84RR364 pKa = 11.84TINVDD369 pKa = 3.2PSVLDD374 pKa = 3.43QLDD377 pKa = 3.89YY378 pKa = 11.61SGDD381 pKa = 3.19WLFTNRR387 pKa = 11.84SGGPVRR393 pKa = 11.84HH394 pKa = 6.39NGFHH398 pKa = 6.54DD399 pKa = 4.16RR400 pKa = 11.84VWRR403 pKa = 11.84PAIKK407 pKa = 10.31RR408 pKa = 11.84ADD410 pKa = 3.71LGVKK414 pKa = 9.82PRR416 pKa = 11.84VHH418 pKa = 7.38DD419 pKa = 4.41LRR421 pKa = 11.84HH422 pKa = 5.77TCASWLIAAGLPLPAIQQHH441 pKa = 6.61LGHH444 pKa = 6.69EE445 pKa = 4.8SIQVTIGVYY454 pKa = 9.36GHH456 pKa = 7.43LDD458 pKa = 3.46RR459 pKa = 11.84SHH461 pKa = 5.77GRR463 pKa = 11.84AVAAAIAAQLDD474 pKa = 4.1RR475 pKa = 11.84NN476 pKa = 3.99
MM1 pKa = 7.0PCYY4 pKa = 10.12RR5 pKa = 11.84RR6 pKa = 11.84THH8 pKa = 5.41GFEE11 pKa = 4.33LRR13 pKa = 11.84ICVGTGARR21 pKa = 11.84GGPRR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84SQQRR31 pKa = 11.84VQSRR35 pKa = 11.84SEE37 pKa = 4.15CVADD41 pKa = 3.0IHH43 pKa = 6.48TPLVVLGIWTFFHH56 pKa = 5.62VVSRR60 pKa = 11.84AVNRR64 pKa = 11.84CFRR67 pKa = 11.84RR68 pKa = 11.84SVPIYY73 pKa = 10.0DD74 pKa = 3.87QISIPGSSTLKK85 pKa = 10.9SPGQRR90 pKa = 11.84HH91 pKa = 5.46FLWPGLFYY99 pKa = 10.75IHH101 pKa = 6.57TPSHH105 pKa = 6.19KK106 pKa = 9.78SLRR109 pKa = 11.84SATMASLRR117 pKa = 11.84TGSRR121 pKa = 11.84KK122 pKa = 10.39DD123 pKa = 3.1GTTYY127 pKa = 8.99TQVRR131 pKa = 11.84YY132 pKa = 9.85RR133 pKa = 11.84LNGKK137 pKa = 7.22QTSTSFDD144 pKa = 3.6DD145 pKa = 3.74PAQAVEE151 pKa = 4.86FKK153 pKa = 11.16RR154 pKa = 11.84MVEE157 pKa = 3.72QLGAAKK163 pKa = 9.9ALEE166 pKa = 4.61VIEE169 pKa = 4.39STEE172 pKa = 3.75ATAQHH177 pKa = 5.66YY178 pKa = 7.57TVGDD182 pKa = 3.64WLDD185 pKa = 3.79HH186 pKa = 5.61YY187 pKa = 10.59LSHH190 pKa = 6.37KK191 pKa = 9.54TGVEE195 pKa = 3.53KK196 pKa = 10.38STMYY200 pKa = 10.58DD201 pKa = 3.23YY202 pKa = 11.63RR203 pKa = 11.84KK204 pKa = 9.14MVEE207 pKa = 3.85KK208 pKa = 10.62DD209 pKa = 2.84IAPVLGPIPLAALTAEE225 pKa = 4.6DD226 pKa = 3.03VAMWVKK232 pKa = 10.63GLAARR237 pKa = 11.84GLKK240 pKa = 10.35GKK242 pKa = 8.65TIANKK247 pKa = 10.32HH248 pKa = 5.45GFLSSALKK256 pKa = 10.03VAASRR261 pKa = 11.84GLIKK265 pKa = 10.8SNPAVGGVGMVEE277 pKa = 4.11IPRR280 pKa = 11.84SEE282 pKa = 4.0RR283 pKa = 11.84EE284 pKa = 3.63EE285 pKa = 4.01MVFLTRR291 pKa = 11.84EE292 pKa = 3.65QYY294 pKa = 11.16ARR296 pKa = 11.84LRR298 pKa = 11.84DD299 pKa = 4.07CVTEE303 pKa = 3.8PWRR306 pKa = 11.84PLVEE310 pKa = 4.14FLVASGARR318 pKa = 11.84WGEE321 pKa = 4.04VTALRR326 pKa = 11.84PSDD329 pKa = 3.63VNRR332 pKa = 11.84DD333 pKa = 3.36EE334 pKa = 4.11GTVRR338 pKa = 11.84ISRR341 pKa = 11.84AWKK344 pKa = 8.46RR345 pKa = 11.84TYY347 pKa = 8.71EE348 pKa = 4.0TGGYY352 pKa = 9.78EE353 pKa = 3.6IGAPKK358 pKa = 9.2TDD360 pKa = 3.04KK361 pKa = 10.73SRR363 pKa = 11.84RR364 pKa = 11.84TINVDD369 pKa = 3.2PSVLDD374 pKa = 3.43QLDD377 pKa = 3.89YY378 pKa = 11.61SGDD381 pKa = 3.19WLFTNRR387 pKa = 11.84SGGPVRR393 pKa = 11.84HH394 pKa = 6.39NGFHH398 pKa = 6.54DD399 pKa = 4.16RR400 pKa = 11.84VWRR403 pKa = 11.84PAIKK407 pKa = 10.31RR408 pKa = 11.84ADD410 pKa = 3.71LGVKK414 pKa = 9.82PRR416 pKa = 11.84VHH418 pKa = 7.38DD419 pKa = 4.41LRR421 pKa = 11.84HH422 pKa = 5.77TCASWLIAAGLPLPAIQQHH441 pKa = 6.61LGHH444 pKa = 6.69EE445 pKa = 4.8SIQVTIGVYY454 pKa = 9.36GHH456 pKa = 7.43LDD458 pKa = 3.46RR459 pKa = 11.84SHH461 pKa = 5.77GRR463 pKa = 11.84AVAAAIAAQLDD474 pKa = 4.1RR475 pKa = 11.84NN476 pKa = 3.99
Molecular weight: 53.05 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
17446 |
39 |
1377 |
223.7 |
24.31 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.696 ± 0.566 | 1.089 ± 0.145 |
6.454 ± 0.255 | 6.087 ± 0.334 |
2.648 ± 0.187 | 8.661 ± 0.532 |
2.218 ± 0.216 | 3.772 ± 0.162 |
3.193 ± 0.226 | 8.294 ± 0.223 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.086 ± 0.117 | 2.786 ± 0.236 |
5.583 ± 0.291 | 3.256 ± 0.194 |
7.549 ± 0.377 | 5.228 ± 0.215 |
5.589 ± 0.195 | 8.34 ± 0.383 |
2.155 ± 0.105 | 2.316 ± 0.198 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
