
Human cyclovirus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; Cyclovirus; unclassified Cyclovirus
Average proteome isoelectric point is 8.7
Get precalculated fractions of proteins
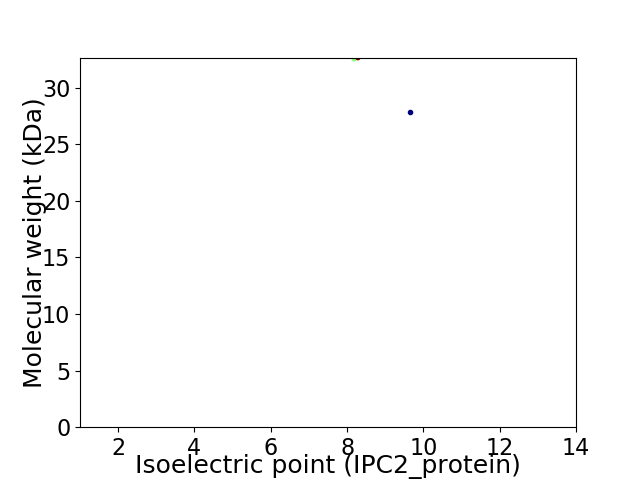
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|W5ZU54|W5ZU54_9CIRC Capsid protein OS=Human cyclovirus OX=1455626 PE=4 SV=1
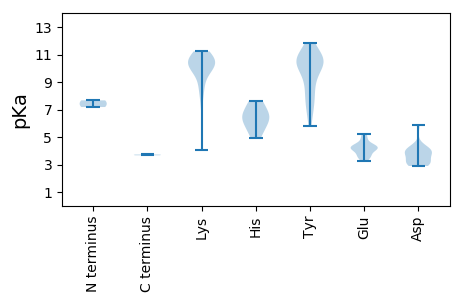
MM1 pKa = 7.18NSVVRR6 pKa = 11.84RR7 pKa = 11.84FCFTWHH13 pKa = 7.63DD14 pKa = 3.96YY15 pKa = 11.15DD16 pKa = 5.87CEE18 pKa = 4.16DD19 pKa = 3.19VAKK22 pKa = 9.79TEE24 pKa = 4.25RR25 pKa = 11.84FINTHH30 pKa = 5.5CKK32 pKa = 9.81YY33 pKa = 10.69GIFGKK38 pKa = 8.63EE39 pKa = 3.59VCPDD43 pKa = 2.94TRR45 pKa = 11.84RR46 pKa = 11.84IHH48 pKa = 6.34LQGFCSLAKK57 pKa = 9.8PKK59 pKa = 10.13RR60 pKa = 11.84FKK62 pKa = 9.89WIKK65 pKa = 8.43EE66 pKa = 3.76QLSNRR71 pKa = 11.84IHH73 pKa = 6.64IEE75 pKa = 3.52KK76 pKa = 10.88AMGSDD81 pKa = 4.36KK82 pKa = 11.06EE83 pKa = 4.14NQQYY87 pKa = 10.51CSKK90 pKa = 10.48SGKK93 pKa = 9.76FFEE96 pKa = 5.1KK97 pKa = 10.55GSPSEE102 pKa = 4.23GSGQRR107 pKa = 11.84TDD109 pKa = 3.0IQSLLEE115 pKa = 4.25TIQGGEE121 pKa = 3.45HH122 pKa = 7.43DD123 pKa = 3.79IRR125 pKa = 11.84RR126 pKa = 11.84IAEE129 pKa = 4.05KK130 pKa = 10.64HH131 pKa = 4.9PACYY135 pKa = 9.3IRR137 pKa = 11.84YY138 pKa = 8.05YY139 pKa = 10.56RR140 pKa = 11.84GIRR143 pKa = 11.84SYY145 pKa = 11.49LNLVAPVSPRR155 pKa = 11.84NFKK158 pKa = 10.36TEE160 pKa = 3.23VRR162 pKa = 11.84YY163 pKa = 9.96YY164 pKa = 9.6WGPPGSGKK172 pKa = 8.77SRR174 pKa = 11.84RR175 pKa = 11.84SLEE178 pKa = 4.08EE179 pKa = 3.49SSGLLDD185 pKa = 3.07GTVYY189 pKa = 10.54YY190 pKa = 10.39KK191 pKa = 10.64PRR193 pKa = 11.84GEE195 pKa = 3.68WWDD198 pKa = 3.98GYY200 pKa = 7.75MQQTSVIIDD209 pKa = 4.09DD210 pKa = 4.39FYY212 pKa = 11.87GWIKK216 pKa = 10.75YY217 pKa = 10.28DD218 pKa = 4.1EE219 pKa = 4.44LLKK222 pKa = 10.69ICDD225 pKa = 4.0RR226 pKa = 11.84YY227 pKa = 8.51PHH229 pKa = 6.56KK230 pKa = 11.23VPIKK234 pKa = 10.44GGFEE238 pKa = 4.18EE239 pKa = 4.92FTSKK243 pKa = 11.17YY244 pKa = 10.6IFITSNVDD252 pKa = 2.89VCDD255 pKa = 4.02LYY257 pKa = 11.53KK258 pKa = 11.06FNGYY262 pKa = 5.83TTAAIDD268 pKa = 3.36RR269 pKa = 11.84RR270 pKa = 11.84ITIKK274 pKa = 10.81EE275 pKa = 3.92NIII278 pKa = 3.67
MM1 pKa = 7.18NSVVRR6 pKa = 11.84RR7 pKa = 11.84FCFTWHH13 pKa = 7.63DD14 pKa = 3.96YY15 pKa = 11.15DD16 pKa = 5.87CEE18 pKa = 4.16DD19 pKa = 3.19VAKK22 pKa = 9.79TEE24 pKa = 4.25RR25 pKa = 11.84FINTHH30 pKa = 5.5CKK32 pKa = 9.81YY33 pKa = 10.69GIFGKK38 pKa = 8.63EE39 pKa = 3.59VCPDD43 pKa = 2.94TRR45 pKa = 11.84RR46 pKa = 11.84IHH48 pKa = 6.34LQGFCSLAKK57 pKa = 9.8PKK59 pKa = 10.13RR60 pKa = 11.84FKK62 pKa = 9.89WIKK65 pKa = 8.43EE66 pKa = 3.76QLSNRR71 pKa = 11.84IHH73 pKa = 6.64IEE75 pKa = 3.52KK76 pKa = 10.88AMGSDD81 pKa = 4.36KK82 pKa = 11.06EE83 pKa = 4.14NQQYY87 pKa = 10.51CSKK90 pKa = 10.48SGKK93 pKa = 9.76FFEE96 pKa = 5.1KK97 pKa = 10.55GSPSEE102 pKa = 4.23GSGQRR107 pKa = 11.84TDD109 pKa = 3.0IQSLLEE115 pKa = 4.25TIQGGEE121 pKa = 3.45HH122 pKa = 7.43DD123 pKa = 3.79IRR125 pKa = 11.84RR126 pKa = 11.84IAEE129 pKa = 4.05KK130 pKa = 10.64HH131 pKa = 4.9PACYY135 pKa = 9.3IRR137 pKa = 11.84YY138 pKa = 8.05YY139 pKa = 10.56RR140 pKa = 11.84GIRR143 pKa = 11.84SYY145 pKa = 11.49LNLVAPVSPRR155 pKa = 11.84NFKK158 pKa = 10.36TEE160 pKa = 3.23VRR162 pKa = 11.84YY163 pKa = 9.96YY164 pKa = 9.6WGPPGSGKK172 pKa = 8.77SRR174 pKa = 11.84RR175 pKa = 11.84SLEE178 pKa = 4.08EE179 pKa = 3.49SSGLLDD185 pKa = 3.07GTVYY189 pKa = 10.54YY190 pKa = 10.39KK191 pKa = 10.64PRR193 pKa = 11.84GEE195 pKa = 3.68WWDD198 pKa = 3.98GYY200 pKa = 7.75MQQTSVIIDD209 pKa = 4.09DD210 pKa = 4.39FYY212 pKa = 11.87GWIKK216 pKa = 10.75YY217 pKa = 10.28DD218 pKa = 4.1EE219 pKa = 4.44LLKK222 pKa = 10.69ICDD225 pKa = 4.0RR226 pKa = 11.84YY227 pKa = 8.51PHH229 pKa = 6.56KK230 pKa = 11.23VPIKK234 pKa = 10.44GGFEE238 pKa = 4.18EE239 pKa = 4.92FTSKK243 pKa = 11.17YY244 pKa = 10.6IFITSNVDD252 pKa = 2.89VCDD255 pKa = 4.02LYY257 pKa = 11.53KK258 pKa = 11.06FNGYY262 pKa = 5.83TTAAIDD268 pKa = 3.36RR269 pKa = 11.84RR270 pKa = 11.84ITIKK274 pKa = 10.81EE275 pKa = 3.92NIII278 pKa = 3.67
Molecular weight: 32.54 kDa
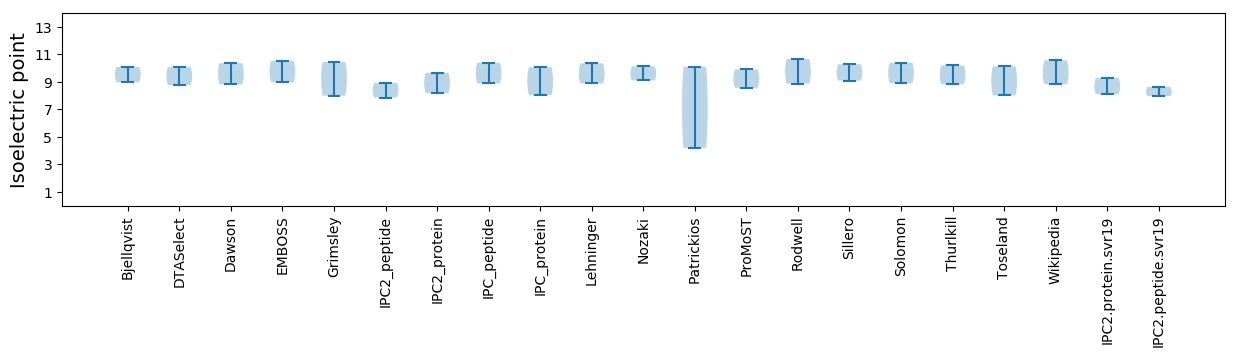
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W5ZU54|W5ZU54_9CIRC Capsid protein OS=Human cyclovirus OX=1455626 PE=4 SV=1
MM1 pKa = 7.67ALRR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84FKK8 pKa = 10.33ARR10 pKa = 11.84RR11 pKa = 11.84PVRR14 pKa = 11.84RR15 pKa = 11.84YY16 pKa = 8.62KK17 pKa = 10.49RR18 pKa = 11.84RR19 pKa = 11.84FMRR22 pKa = 11.84YY23 pKa = 6.66RR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84KK28 pKa = 8.04YY29 pKa = 7.41QQRR32 pKa = 11.84VKK34 pKa = 9.76PGDD37 pKa = 3.54MIIQLKK43 pKa = 10.19RR44 pKa = 11.84IRR46 pKa = 11.84TVKK49 pKa = 10.47VNSQVNEE56 pKa = 4.08YY57 pKa = 10.4IGLNFTAKK65 pKa = 10.32DD66 pKa = 3.97FPEE69 pKa = 4.44LQPFLTQFEE78 pKa = 4.37AFQFQRR84 pKa = 11.84YY85 pKa = 6.89AVKK88 pKa = 10.56VIPSMNVSNNSTSRR102 pKa = 11.84IGTYY106 pKa = 10.59CLVPWHH112 pKa = 6.89KK113 pKa = 10.2KK114 pKa = 4.05TTKK117 pKa = 7.32PTNYY121 pKa = 10.33SSMLSIDD128 pKa = 3.08KK129 pKa = 10.74SRR131 pKa = 11.84VYY133 pKa = 10.59RR134 pKa = 11.84GTQTAYY140 pKa = 10.54RR141 pKa = 11.84SFVPAVHH148 pKa = 5.76VQAEE152 pKa = 4.48TTGTGDD158 pKa = 3.11AAAVIKK164 pKa = 8.99WKK166 pKa = 10.01PRR168 pKa = 11.84IDD170 pKa = 3.76IVNLYY175 pKa = 10.51NDD177 pKa = 3.35VEE179 pKa = 5.23FYY181 pKa = 11.09CGMIGIQGIPDD192 pKa = 3.04ATEE195 pKa = 3.64NVNYY199 pKa = 9.14YY200 pKa = 10.45HH201 pKa = 6.11VVEE204 pKa = 4.39YY205 pKa = 7.72VTCKK209 pKa = 10.69LINQNYY215 pKa = 9.48LNDD218 pKa = 3.77KK219 pKa = 10.1AVQARR224 pKa = 11.84ALEE227 pKa = 4.33EE228 pKa = 4.24SFSEE232 pKa = 4.18AFEE235 pKa = 4.84AII237 pKa = 3.73
MM1 pKa = 7.67ALRR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84FKK8 pKa = 10.33ARR10 pKa = 11.84RR11 pKa = 11.84PVRR14 pKa = 11.84RR15 pKa = 11.84YY16 pKa = 8.62KK17 pKa = 10.49RR18 pKa = 11.84RR19 pKa = 11.84FMRR22 pKa = 11.84YY23 pKa = 6.66RR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84KK28 pKa = 8.04YY29 pKa = 7.41QQRR32 pKa = 11.84VKK34 pKa = 9.76PGDD37 pKa = 3.54MIIQLKK43 pKa = 10.19RR44 pKa = 11.84IRR46 pKa = 11.84TVKK49 pKa = 10.47VNSQVNEE56 pKa = 4.08YY57 pKa = 10.4IGLNFTAKK65 pKa = 10.32DD66 pKa = 3.97FPEE69 pKa = 4.44LQPFLTQFEE78 pKa = 4.37AFQFQRR84 pKa = 11.84YY85 pKa = 6.89AVKK88 pKa = 10.56VIPSMNVSNNSTSRR102 pKa = 11.84IGTYY106 pKa = 10.59CLVPWHH112 pKa = 6.89KK113 pKa = 10.2KK114 pKa = 4.05TTKK117 pKa = 7.32PTNYY121 pKa = 10.33SSMLSIDD128 pKa = 3.08KK129 pKa = 10.74SRR131 pKa = 11.84VYY133 pKa = 10.59RR134 pKa = 11.84GTQTAYY140 pKa = 10.54RR141 pKa = 11.84SFVPAVHH148 pKa = 5.76VQAEE152 pKa = 4.48TTGTGDD158 pKa = 3.11AAAVIKK164 pKa = 8.99WKK166 pKa = 10.01PRR168 pKa = 11.84IDD170 pKa = 3.76IVNLYY175 pKa = 10.51NDD177 pKa = 3.35VEE179 pKa = 5.23FYY181 pKa = 11.09CGMIGIQGIPDD192 pKa = 3.04ATEE195 pKa = 3.64NVNYY199 pKa = 9.14YY200 pKa = 10.45HH201 pKa = 6.11VVEE204 pKa = 4.39YY205 pKa = 7.72VTCKK209 pKa = 10.69LINQNYY215 pKa = 9.48LNDD218 pKa = 3.77KK219 pKa = 10.1AVQARR224 pKa = 11.84ALEE227 pKa = 4.33EE228 pKa = 4.24SFSEE232 pKa = 4.18AFEE235 pKa = 4.84AII237 pKa = 3.73
Molecular weight: 27.79 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
515 |
237 |
278 |
257.5 |
30.17 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.854 ± 1.397 | 2.33 ± 0.641 |
4.66 ± 0.774 | 5.825 ± 0.713 |
5.049 ± 0.009 | 6.019 ± 1.339 |
1.942 ± 0.407 | 7.573 ± 0.749 |
7.573 ± 0.495 | 4.66 ± 0.011 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.748 ± 0.472 | 4.466 ± 0.868 |
4.078 ± 0.085 | 4.272 ± 0.731 |
8.738 ± 0.837 | 6.214 ± 0.693 |
5.631 ± 0.421 | 6.408 ± 1.478 |
1.553 ± 0.427 | 6.408 ± 0.047 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
