
Chilo suppressalis (Asiatic rice borer moth)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Protostomia; Ecdysozoa; Panarthropoda; Arthropoda; Mandibulata; Pancrustacea; Hexapoda; Insecta; Dicondylia; Pterygota; Neoptera; Endopterygota; Amphiesmenoptera; Lepidoptera; Glossata;
Average proteome isoelectric point is 6.98
Get precalculated fractions of proteins
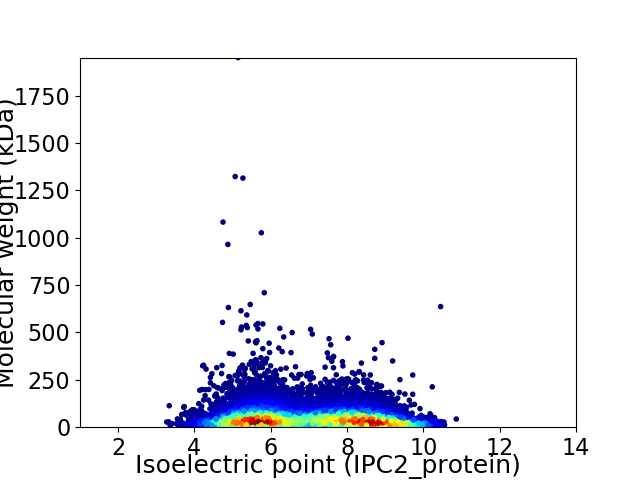
Virtual 2D-PAGE plot for 15558 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3S2NGZ1|A0A3S2NGZ1_CHISP ABC transporter domain-containing protein (Fragment) OS=Chilo suppressalis OX=168631 GN=evm_015542 PE=4 SV=1
MM1 pKa = 7.48CRR3 pKa = 11.84NSSFSSILDD12 pKa = 3.48LALILDD18 pKa = 4.38NNDD21 pKa = 3.56NDD23 pKa = 5.0DD24 pKa = 3.74STSNRR29 pKa = 11.84NTHH32 pKa = 6.76DD33 pKa = 3.6EE34 pKa = 4.13NSNLEE39 pKa = 4.24PDD41 pKa = 4.57DD42 pKa = 4.53PCDD45 pKa = 3.31TAMSSPLPPGARR57 pKa = 11.84NIKK60 pKa = 10.25EE61 pKa = 3.99DD62 pKa = 3.6MAEE65 pKa = 3.72VWNDD69 pKa = 3.16VFNNNYY75 pKa = 9.97PIDD78 pKa = 4.12FSQYY82 pKa = 9.6QLQATYY88 pKa = 8.43DD89 pKa = 3.56TTFPCC94 pKa = 5.41
MM1 pKa = 7.48CRR3 pKa = 11.84NSSFSSILDD12 pKa = 3.48LALILDD18 pKa = 4.38NNDD21 pKa = 3.56NDD23 pKa = 5.0DD24 pKa = 3.74STSNRR29 pKa = 11.84NTHH32 pKa = 6.76DD33 pKa = 3.6EE34 pKa = 4.13NSNLEE39 pKa = 4.24PDD41 pKa = 4.57DD42 pKa = 4.53PCDD45 pKa = 3.31TAMSSPLPPGARR57 pKa = 11.84NIKK60 pKa = 10.25EE61 pKa = 3.99DD62 pKa = 3.6MAEE65 pKa = 3.72VWNDD69 pKa = 3.16VFNNNYY75 pKa = 9.97PIDD78 pKa = 4.12FSQYY82 pKa = 9.6QLQATYY88 pKa = 8.43DD89 pKa = 3.56TTFPCC94 pKa = 5.41
Molecular weight: 10.61 kDa
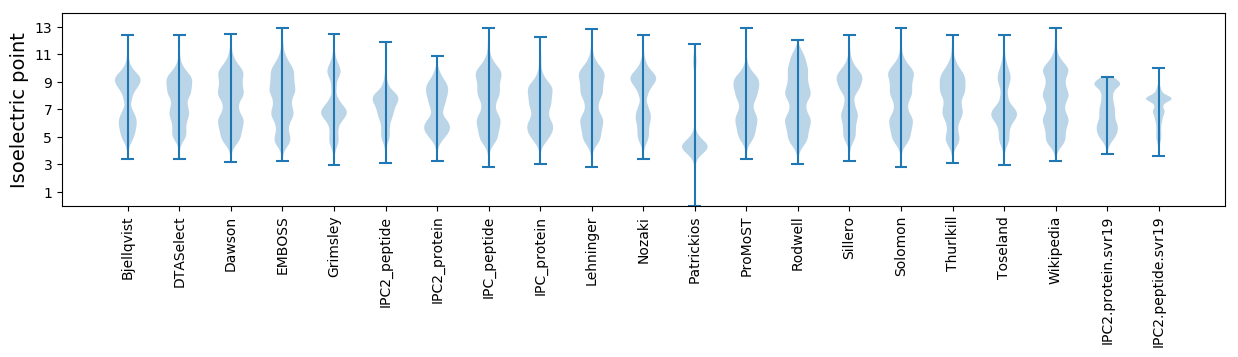
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A437BMJ8|A0A437BMJ8_CHISP Uncharacterized protein (Fragment) OS=Chilo suppressalis OX=168631 GN=evm_003678 PE=4 SV=1
MM1 pKa = 7.18PQNGLRR7 pKa = 11.84GRR9 pKa = 11.84GTEE12 pKa = 3.85IEE14 pKa = 4.23GLAVAVAVATSMQSFVVLRR33 pKa = 11.84CDD35 pKa = 3.82CVIDD39 pKa = 4.81LLVLLWAHH47 pKa = 5.9NQLKK51 pKa = 9.78LAKK54 pKa = 9.64SYY56 pKa = 11.48YY57 pKa = 10.63GLLKK61 pKa = 10.82DD62 pKa = 4.18GIGRR66 pKa = 11.84LGHH69 pKa = 6.65DD70 pKa = 3.85PPRR73 pKa = 11.84GLSADD78 pKa = 2.84WWVLTTADD86 pKa = 3.41AAGTNGLTSGGFVVIATLGRR106 pKa = 11.84RR107 pKa = 11.84VGDD110 pKa = 3.12RR111 pKa = 11.84SVAMEE116 pKa = 4.5LKK118 pKa = 10.22EE119 pKa = 4.03KK120 pKa = 9.99TLLSVMTPMTPPLRR134 pKa = 11.84LLRR137 pKa = 11.84HH138 pKa = 5.92PWDD141 pKa = 3.67KK142 pKa = 11.82GEE144 pKa = 3.76WCYY147 pKa = 11.59SRR149 pKa = 11.84LEE151 pKa = 4.15HH152 pKa = 6.06TRR154 pKa = 11.84HH155 pKa = 5.49FFVKK159 pKa = 10.55FMLITTQSNLRR170 pKa = 11.84IFTSIACAFPNLRR183 pKa = 11.84GTNIRR188 pKa = 11.84TSPLRR193 pKa = 11.84VRR195 pKa = 11.84KK196 pKa = 9.97LIRR199 pKa = 11.84CHH201 pKa = 6.49SVSTKK206 pKa = 9.91LVRR209 pKa = 11.84RR210 pKa = 11.84AQKK213 pKa = 9.52TEE215 pKa = 3.84TPHH218 pKa = 6.76PISYY222 pKa = 9.36MIFLEE227 pKa = 4.11NWQKK231 pKa = 10.91RR232 pKa = 11.84ALLLRR237 pKa = 11.84LYY239 pKa = 10.49CGVAPCIGGCSRR251 pKa = 11.84DD252 pKa = 3.88DD253 pKa = 3.67FTKK256 pKa = 10.52KK257 pKa = 10.25SYY259 pKa = 10.5IGIGVAIQILPGPHH273 pKa = 5.52RR274 pKa = 11.84TLVTEE279 pKa = 4.47GTLLSHH285 pKa = 7.08TLTISVARR293 pKa = 11.84AAGLPHH299 pKa = 6.55TVLHH303 pKa = 6.39HH304 pKa = 5.3QLAVPVRR311 pKa = 11.84LVRR314 pKa = 11.84RR315 pKa = 11.84TPEE318 pKa = 4.75DD319 pKa = 3.04IRR321 pKa = 11.84KK322 pKa = 9.71SSPILRR328 pKa = 11.84RR329 pKa = 11.84AEE331 pKa = 4.04RR332 pKa = 11.84VDD334 pKa = 3.28TDD336 pKa = 3.58VVRR339 pKa = 11.84ARR341 pKa = 11.84LLRR344 pKa = 11.84EE345 pKa = 3.86LSGEE349 pKa = 4.05ALVGAVQAAALARR362 pKa = 11.84HH363 pKa = 6.33AGALHH368 pKa = 6.99LSRR371 pKa = 11.84HH372 pKa = 5.12SRR374 pKa = 11.84SGRR377 pKa = 11.84DD378 pKa = 3.34GTADD382 pKa = 3.21AAQYY386 pKa = 10.51RR387 pKa = 11.84AVHH390 pKa = 6.52DD391 pKa = 4.16PGIRR395 pKa = 11.84DD396 pKa = 3.47HH397 pKa = 6.62VAAGVAASQAPLPNVAACRR416 pKa = 11.84RR417 pKa = 11.84LL418 pKa = 3.29
MM1 pKa = 7.18PQNGLRR7 pKa = 11.84GRR9 pKa = 11.84GTEE12 pKa = 3.85IEE14 pKa = 4.23GLAVAVAVATSMQSFVVLRR33 pKa = 11.84CDD35 pKa = 3.82CVIDD39 pKa = 4.81LLVLLWAHH47 pKa = 5.9NQLKK51 pKa = 9.78LAKK54 pKa = 9.64SYY56 pKa = 11.48YY57 pKa = 10.63GLLKK61 pKa = 10.82DD62 pKa = 4.18GIGRR66 pKa = 11.84LGHH69 pKa = 6.65DD70 pKa = 3.85PPRR73 pKa = 11.84GLSADD78 pKa = 2.84WWVLTTADD86 pKa = 3.41AAGTNGLTSGGFVVIATLGRR106 pKa = 11.84RR107 pKa = 11.84VGDD110 pKa = 3.12RR111 pKa = 11.84SVAMEE116 pKa = 4.5LKK118 pKa = 10.22EE119 pKa = 4.03KK120 pKa = 9.99TLLSVMTPMTPPLRR134 pKa = 11.84LLRR137 pKa = 11.84HH138 pKa = 5.92PWDD141 pKa = 3.67KK142 pKa = 11.82GEE144 pKa = 3.76WCYY147 pKa = 11.59SRR149 pKa = 11.84LEE151 pKa = 4.15HH152 pKa = 6.06TRR154 pKa = 11.84HH155 pKa = 5.49FFVKK159 pKa = 10.55FMLITTQSNLRR170 pKa = 11.84IFTSIACAFPNLRR183 pKa = 11.84GTNIRR188 pKa = 11.84TSPLRR193 pKa = 11.84VRR195 pKa = 11.84KK196 pKa = 9.97LIRR199 pKa = 11.84CHH201 pKa = 6.49SVSTKK206 pKa = 9.91LVRR209 pKa = 11.84RR210 pKa = 11.84AQKK213 pKa = 9.52TEE215 pKa = 3.84TPHH218 pKa = 6.76PISYY222 pKa = 9.36MIFLEE227 pKa = 4.11NWQKK231 pKa = 10.91RR232 pKa = 11.84ALLLRR237 pKa = 11.84LYY239 pKa = 10.49CGVAPCIGGCSRR251 pKa = 11.84DD252 pKa = 3.88DD253 pKa = 3.67FTKK256 pKa = 10.52KK257 pKa = 10.25SYY259 pKa = 10.5IGIGVAIQILPGPHH273 pKa = 5.52RR274 pKa = 11.84TLVTEE279 pKa = 4.47GTLLSHH285 pKa = 7.08TLTISVARR293 pKa = 11.84AAGLPHH299 pKa = 6.55TVLHH303 pKa = 6.39HH304 pKa = 5.3QLAVPVRR311 pKa = 11.84LVRR314 pKa = 11.84RR315 pKa = 11.84TPEE318 pKa = 4.75DD319 pKa = 3.04IRR321 pKa = 11.84KK322 pKa = 9.71SSPILRR328 pKa = 11.84RR329 pKa = 11.84AEE331 pKa = 4.04RR332 pKa = 11.84VDD334 pKa = 3.28TDD336 pKa = 3.58VVRR339 pKa = 11.84ARR341 pKa = 11.84LLRR344 pKa = 11.84EE345 pKa = 3.86LSGEE349 pKa = 4.05ALVGAVQAAALARR362 pKa = 11.84HH363 pKa = 6.33AGALHH368 pKa = 6.99LSRR371 pKa = 11.84HH372 pKa = 5.12SRR374 pKa = 11.84SGRR377 pKa = 11.84DD378 pKa = 3.34GTADD382 pKa = 3.21AAQYY386 pKa = 10.51RR387 pKa = 11.84AVHH390 pKa = 6.52DD391 pKa = 4.16PGIRR395 pKa = 11.84DD396 pKa = 3.47HH397 pKa = 6.62VAAGVAASQAPLPNVAACRR416 pKa = 11.84RR417 pKa = 11.84LL418 pKa = 3.29
Molecular weight: 45.91 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
7277707 |
52 |
18336 |
467.8 |
52.51 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.107 ± 0.03 | 2.131 ± 0.036 |
5.474 ± 0.016 | 6.481 ± 0.03 |
3.508 ± 0.016 | 5.602 ± 0.028 |
2.584 ± 0.013 | 5.294 ± 0.021 |
6.315 ± 0.032 | 9.067 ± 0.029 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.227 ± 0.01 | 4.828 ± 0.023 |
5.549 ± 0.033 | 3.912 ± 0.02 |
5.833 ± 0.025 | 7.674 ± 0.022 |
5.796 ± 0.023 | 6.297 ± 0.015 |
1.14 ± 0.008 | 3.18 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
