
Pea stem necrosis virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Tolucaviricetes; Tolivirales; Tombusviridae; Procedovirinae; Gammacarmovirus
Average proteome isoelectric point is 7.87
Get precalculated fractions of proteins
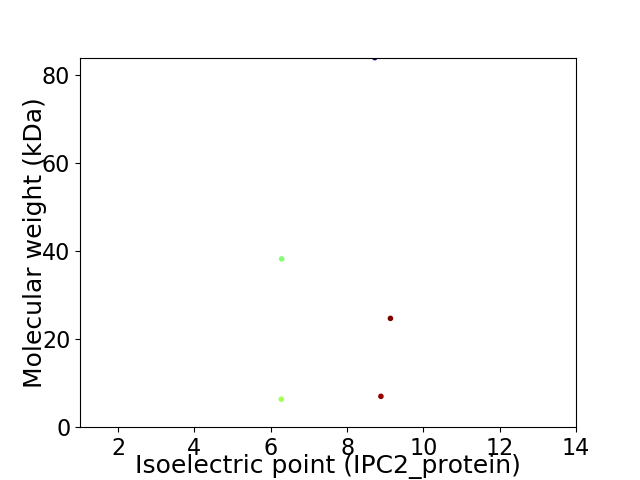
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
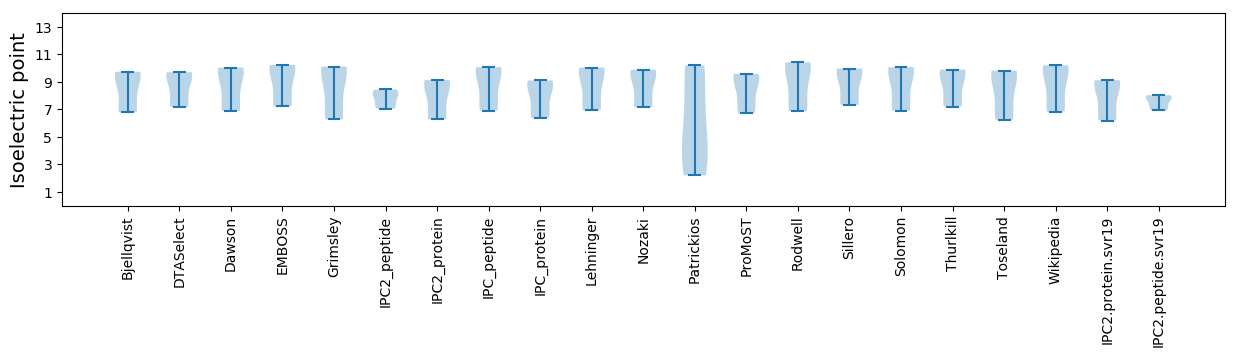
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q7T4K0|Q7T4K0_9TOMB p6 product OS=Pea stem necrosis virus OX=199361 GN=p6 PE=4 SV=1
MM1 pKa = 7.6GNLPNNSDD9 pKa = 4.07DD10 pKa = 3.45FWVNKK15 pKa = 8.67VRR17 pKa = 11.84GWWRR21 pKa = 11.84RR22 pKa = 11.84ASRR25 pKa = 11.84EE26 pKa = 4.24SYY28 pKa = 10.6SSCGCCDD35 pKa = 3.25RR36 pKa = 11.84CPWKK40 pKa = 10.7APCYY44 pKa = 9.89HH45 pKa = 6.39SRR47 pKa = 11.84NSNTVVTHH55 pKa = 6.29RR56 pKa = 11.84EE57 pKa = 3.93YY58 pKa = 10.77VTDD61 pKa = 3.62VSNSSGVFVNNSSGSSSVFRR81 pKa = 11.84INPSNTRR88 pKa = 11.84VFNWLSSIAGSYY100 pKa = 10.83DD101 pKa = 3.11SYY103 pKa = 11.54KK104 pKa = 8.89FTRR107 pKa = 11.84VRR109 pKa = 11.84FTYY112 pKa = 10.41VPYY115 pKa = 10.63AGSNTPGRR123 pKa = 11.84LYY125 pKa = 10.81LGWDD129 pKa = 3.55ADD131 pKa = 4.2SQDD134 pKa = 3.69VIQPDD139 pKa = 3.79RR140 pKa = 11.84ASLANFSPTSEE151 pKa = 4.07DD152 pKa = 3.91SVWTDD157 pKa = 2.7SYY159 pKa = 11.8LVIPVDD165 pKa = 3.82RR166 pKa = 11.84EE167 pKa = 3.73WRR169 pKa = 11.84FVDD172 pKa = 3.4DD173 pKa = 4.04TNISSRR179 pKa = 11.84KK180 pKa = 9.68LVDD183 pKa = 4.02LGQLVLATWGGVDD196 pKa = 3.2NTVCGEE202 pKa = 4.18VYY204 pKa = 10.38VEE206 pKa = 3.9YY207 pKa = 10.46SVEE210 pKa = 3.79LRR212 pKa = 11.84QPQPPSGFVQVGRR225 pKa = 11.84VDD227 pKa = 3.33VPGILTFTGPAFVPASDD244 pKa = 4.96LSISDD249 pKa = 3.34TSFSFNLNTAGQYY262 pKa = 10.97ALSIDD267 pKa = 4.02LQATTSGTLTVAGNCTLLGVVKK289 pKa = 10.44SQFASGVGIYY299 pKa = 9.05MCIVRR304 pKa = 11.84STGGPNSAASISIGTLTGLSRR325 pKa = 11.84VQFFLSRR332 pKa = 11.84GTTSAGIRR340 pKa = 11.84VTCISSEE347 pKa = 3.97RR348 pKa = 11.84HH349 pKa = 5.2LSS351 pKa = 3.4
MM1 pKa = 7.6GNLPNNSDD9 pKa = 4.07DD10 pKa = 3.45FWVNKK15 pKa = 8.67VRR17 pKa = 11.84GWWRR21 pKa = 11.84RR22 pKa = 11.84ASRR25 pKa = 11.84EE26 pKa = 4.24SYY28 pKa = 10.6SSCGCCDD35 pKa = 3.25RR36 pKa = 11.84CPWKK40 pKa = 10.7APCYY44 pKa = 9.89HH45 pKa = 6.39SRR47 pKa = 11.84NSNTVVTHH55 pKa = 6.29RR56 pKa = 11.84EE57 pKa = 3.93YY58 pKa = 10.77VTDD61 pKa = 3.62VSNSSGVFVNNSSGSSSVFRR81 pKa = 11.84INPSNTRR88 pKa = 11.84VFNWLSSIAGSYY100 pKa = 10.83DD101 pKa = 3.11SYY103 pKa = 11.54KK104 pKa = 8.89FTRR107 pKa = 11.84VRR109 pKa = 11.84FTYY112 pKa = 10.41VPYY115 pKa = 10.63AGSNTPGRR123 pKa = 11.84LYY125 pKa = 10.81LGWDD129 pKa = 3.55ADD131 pKa = 4.2SQDD134 pKa = 3.69VIQPDD139 pKa = 3.79RR140 pKa = 11.84ASLANFSPTSEE151 pKa = 4.07DD152 pKa = 3.91SVWTDD157 pKa = 2.7SYY159 pKa = 11.8LVIPVDD165 pKa = 3.82RR166 pKa = 11.84EE167 pKa = 3.73WRR169 pKa = 11.84FVDD172 pKa = 3.4DD173 pKa = 4.04TNISSRR179 pKa = 11.84KK180 pKa = 9.68LVDD183 pKa = 4.02LGQLVLATWGGVDD196 pKa = 3.2NTVCGEE202 pKa = 4.18VYY204 pKa = 10.38VEE206 pKa = 3.9YY207 pKa = 10.46SVEE210 pKa = 3.79LRR212 pKa = 11.84QPQPPSGFVQVGRR225 pKa = 11.84VDD227 pKa = 3.33VPGILTFTGPAFVPASDD244 pKa = 4.96LSISDD249 pKa = 3.34TSFSFNLNTAGQYY262 pKa = 10.97ALSIDD267 pKa = 4.02LQATTSGTLTVAGNCTLLGVVKK289 pKa = 10.44SQFASGVGIYY299 pKa = 9.05MCIVRR304 pKa = 11.84STGGPNSAASISIGTLTGLSRR325 pKa = 11.84VQFFLSRR332 pKa = 11.84GTTSAGIRR340 pKa = 11.84VTCISSEE347 pKa = 3.97RR348 pKa = 11.84HH349 pKa = 5.2LSS351 pKa = 3.4
Molecular weight: 38.23 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q7T4K3|Q7T4K3_9TOMB RNA-directed RNA polymerase OS=Pea stem necrosis virus OX=199361 GN=p84 PE=4 SV=2
MM1 pKa = 7.67SIRR4 pKa = 11.84MKK6 pKa = 10.79FGLALVAGCASVMYY20 pKa = 9.11VAYY23 pKa = 10.0RR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84VVRR29 pKa = 11.84GKK31 pKa = 9.11MLPRR35 pKa = 11.84NLQIEE40 pKa = 4.29NSRR43 pKa = 11.84VVLKK47 pKa = 10.64VLNSQSDD54 pKa = 3.97GVDD57 pKa = 3.93FPEE60 pKa = 4.66EE61 pKa = 4.13DD62 pKa = 4.0SVDD65 pKa = 3.39SSGPVILIGSVEE77 pKa = 4.46VKK79 pKa = 10.54LPAPVTVKK87 pKa = 10.5RR88 pKa = 11.84KK89 pKa = 9.46PKK91 pKa = 9.99AKK93 pKa = 9.39HH94 pKa = 5.83KK95 pKa = 10.73SMPFITKK102 pKa = 10.14LINAAKK108 pKa = 9.76VHH110 pKa = 6.44FDD112 pKa = 3.53GVPKK116 pKa = 9.16PTEE119 pKa = 4.24SNHH122 pKa = 4.85MAVTRR127 pKa = 11.84FIKK130 pKa = 10.06DD131 pKa = 3.13YY132 pKa = 10.31CKK134 pKa = 10.66EE135 pKa = 4.0HH136 pKa = 7.01GVDD139 pKa = 3.91DD140 pKa = 4.04NQTRR144 pKa = 11.84RR145 pKa = 11.84VCAIAGPLILSPDD158 pKa = 3.26RR159 pKa = 11.84TDD161 pKa = 2.76IMSRR165 pKa = 11.84AFLYY169 pKa = 10.81GPEE172 pKa = 4.22LSKK175 pKa = 10.95QRR177 pKa = 11.84AEE179 pKa = 4.06YY180 pKa = 10.1AAAASTISWFDD191 pKa = 4.26AIVAAPLSCRR201 pKa = 11.84AWRR204 pKa = 11.84RR205 pKa = 11.84AYY207 pKa = 9.98LALMGYY213 pKa = 8.4PDD215 pKa = 3.59SVGYY219 pKa = 10.55QMVKK223 pKa = 10.58
MM1 pKa = 7.67SIRR4 pKa = 11.84MKK6 pKa = 10.79FGLALVAGCASVMYY20 pKa = 9.11VAYY23 pKa = 10.0RR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84VVRR29 pKa = 11.84GKK31 pKa = 9.11MLPRR35 pKa = 11.84NLQIEE40 pKa = 4.29NSRR43 pKa = 11.84VVLKK47 pKa = 10.64VLNSQSDD54 pKa = 3.97GVDD57 pKa = 3.93FPEE60 pKa = 4.66EE61 pKa = 4.13DD62 pKa = 4.0SVDD65 pKa = 3.39SSGPVILIGSVEE77 pKa = 4.46VKK79 pKa = 10.54LPAPVTVKK87 pKa = 10.5RR88 pKa = 11.84KK89 pKa = 9.46PKK91 pKa = 9.99AKK93 pKa = 9.39HH94 pKa = 5.83KK95 pKa = 10.73SMPFITKK102 pKa = 10.14LINAAKK108 pKa = 9.76VHH110 pKa = 6.44FDD112 pKa = 3.53GVPKK116 pKa = 9.16PTEE119 pKa = 4.24SNHH122 pKa = 4.85MAVTRR127 pKa = 11.84FIKK130 pKa = 10.06DD131 pKa = 3.13YY132 pKa = 10.31CKK134 pKa = 10.66EE135 pKa = 4.0HH136 pKa = 7.01GVDD139 pKa = 3.91DD140 pKa = 4.04NQTRR144 pKa = 11.84RR145 pKa = 11.84VCAIAGPLILSPDD158 pKa = 3.26RR159 pKa = 11.84TDD161 pKa = 2.76IMSRR165 pKa = 11.84AFLYY169 pKa = 10.81GPEE172 pKa = 4.22LSKK175 pKa = 10.95QRR177 pKa = 11.84AEE179 pKa = 4.06YY180 pKa = 10.1AAAASTISWFDD191 pKa = 4.26AIVAAPLSCRR201 pKa = 11.84AWRR204 pKa = 11.84RR205 pKa = 11.84AYY207 pKa = 9.98LALMGYY213 pKa = 8.4PDD215 pKa = 3.59SVGYY219 pKa = 10.55QMVKK223 pKa = 10.58
Molecular weight: 24.72 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1444 |
57 |
750 |
288.8 |
32.05 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.202 ± 0.985 | 2.424 ± 0.328 |
4.778 ± 0.375 | 4.155 ± 0.736 |
4.294 ± 0.395 | 7.202 ± 0.591 |
2.008 ± 0.446 | 5.471 ± 0.658 |
5.194 ± 1.306 | 7.825 ± 0.889 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.355 ± 0.78 | 4.432 ± 0.662 |
4.848 ± 0.397 | 3.324 ± 0.347 |
6.925 ± 0.319 | 9.418 ± 1.689 |
4.224 ± 1.148 | 9.003 ± 0.562 |
1.385 ± 0.375 | 3.463 ± 0.092 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
