
Winogradskyella epiphytica
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Winogradskyella
Average proteome isoelectric point is 6.42
Get precalculated fractions of proteins
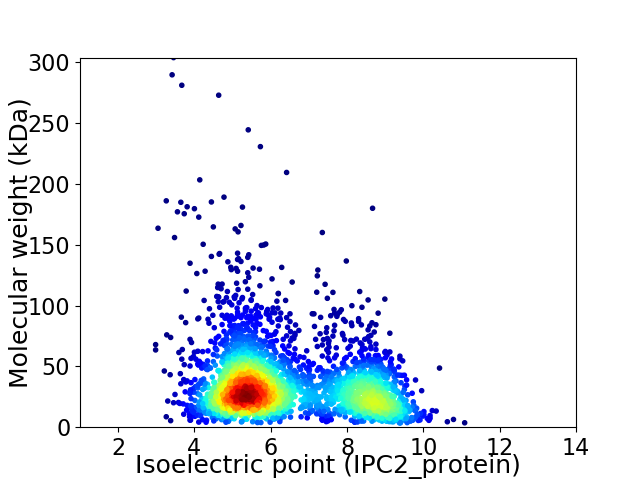
Virtual 2D-PAGE plot for 3007 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2V4WV60|A0A2V4WV60_9FLAO Uncharacterized protein OS=Winogradskyella epiphytica OX=262005 GN=DFQ11_10466 PE=4 SV=1
MM1 pKa = 7.5KK2 pKa = 10.55LKK4 pKa = 10.44FLRR7 pKa = 11.84LSLLLLAVFSVVISCKK23 pKa = 10.49DD24 pKa = 3.22DD25 pKa = 4.4DD26 pKa = 4.78DD27 pKa = 5.22FEE29 pKa = 5.15EE30 pKa = 4.81IPYY33 pKa = 10.22VLEE36 pKa = 5.69DD37 pKa = 3.49RR38 pKa = 11.84TEE40 pKa = 3.9QQAKK44 pKa = 10.65DD45 pKa = 3.55KK46 pKa = 11.23DD47 pKa = 3.93SLLSYY52 pKa = 10.94LSTHH56 pKa = 6.63YY57 pKa = 10.77YY58 pKa = 10.31NSAFFEE64 pKa = 4.39TGTNHH69 pKa = 7.44RR70 pKa = 11.84YY71 pKa = 9.15NDD73 pKa = 3.4IVITEE78 pKa = 4.25LPKK81 pKa = 10.96DD82 pKa = 3.46EE83 pKa = 4.62SGNYY87 pKa = 10.34LEE89 pKa = 5.88MPDD92 pKa = 4.43PEE94 pKa = 4.93NNTLLIEE101 pKa = 4.29AVEE104 pKa = 4.3TLTTEE109 pKa = 4.26YY110 pKa = 11.07KK111 pKa = 10.36DD112 pKa = 3.32VSYY115 pKa = 10.55EE116 pKa = 4.1YY117 pKa = 11.06YY118 pKa = 10.73VLNINQGGGEE128 pKa = 4.38SPNFTDD134 pKa = 3.66KK135 pKa = 11.04VRR137 pKa = 11.84VRR139 pKa = 11.84YY140 pKa = 9.93QGVSVGNQTVFDD152 pKa = 4.32AVSSPITLPLVGDD165 pKa = 4.5GFQTGGAIRR174 pKa = 11.84AWQLVIPTFKK184 pKa = 10.22TAVSYY189 pKa = 10.79SYY191 pKa = 11.38NDD193 pKa = 3.56GVVEE197 pKa = 3.96YY198 pKa = 10.58DD199 pKa = 3.79DD200 pKa = 5.64FGLGMMFVPSGLAYY214 pKa = 10.13FAGVNTGSAYY224 pKa = 10.83DD225 pKa = 3.52NLIFKK230 pKa = 10.62FEE232 pKa = 4.22LLQYY236 pKa = 10.36EE237 pKa = 4.77VVDD240 pKa = 4.35HH241 pKa = 7.41DD242 pKa = 4.9SDD244 pKa = 5.61GIPSWVEE251 pKa = 3.89DD252 pKa = 3.97LDD254 pKa = 5.86GDD256 pKa = 4.91LDD258 pKa = 4.15VSNDD262 pKa = 3.52DD263 pKa = 3.38TDD265 pKa = 5.06GNGFPNYY272 pKa = 9.79VDD274 pKa = 5.0RR275 pKa = 11.84DD276 pKa = 3.85DD277 pKa = 6.18DD278 pKa = 4.75GDD280 pKa = 4.05GVLTRR285 pKa = 11.84DD286 pKa = 3.45EE287 pKa = 5.71LIPTTYY293 pKa = 10.44TIDD296 pKa = 3.54LNAGEE301 pKa = 4.67EE302 pKa = 4.34EE303 pKa = 4.39PVLSEE308 pKa = 4.63GEE310 pKa = 4.3FEE312 pKa = 5.11VSRR315 pKa = 11.84DD316 pKa = 3.67LNNGILTIEE325 pKa = 4.37TVTIADD331 pKa = 3.96SNNDD335 pKa = 3.08GLADD339 pKa = 3.76YY340 pKa = 10.31LDD342 pKa = 4.17PNITINYY349 pKa = 9.07NEE351 pKa = 3.96
MM1 pKa = 7.5KK2 pKa = 10.55LKK4 pKa = 10.44FLRR7 pKa = 11.84LSLLLLAVFSVVISCKK23 pKa = 10.49DD24 pKa = 3.22DD25 pKa = 4.4DD26 pKa = 4.78DD27 pKa = 5.22FEE29 pKa = 5.15EE30 pKa = 4.81IPYY33 pKa = 10.22VLEE36 pKa = 5.69DD37 pKa = 3.49RR38 pKa = 11.84TEE40 pKa = 3.9QQAKK44 pKa = 10.65DD45 pKa = 3.55KK46 pKa = 11.23DD47 pKa = 3.93SLLSYY52 pKa = 10.94LSTHH56 pKa = 6.63YY57 pKa = 10.77YY58 pKa = 10.31NSAFFEE64 pKa = 4.39TGTNHH69 pKa = 7.44RR70 pKa = 11.84YY71 pKa = 9.15NDD73 pKa = 3.4IVITEE78 pKa = 4.25LPKK81 pKa = 10.96DD82 pKa = 3.46EE83 pKa = 4.62SGNYY87 pKa = 10.34LEE89 pKa = 5.88MPDD92 pKa = 4.43PEE94 pKa = 4.93NNTLLIEE101 pKa = 4.29AVEE104 pKa = 4.3TLTTEE109 pKa = 4.26YY110 pKa = 11.07KK111 pKa = 10.36DD112 pKa = 3.32VSYY115 pKa = 10.55EE116 pKa = 4.1YY117 pKa = 11.06YY118 pKa = 10.73VLNINQGGGEE128 pKa = 4.38SPNFTDD134 pKa = 3.66KK135 pKa = 11.04VRR137 pKa = 11.84VRR139 pKa = 11.84YY140 pKa = 9.93QGVSVGNQTVFDD152 pKa = 4.32AVSSPITLPLVGDD165 pKa = 4.5GFQTGGAIRR174 pKa = 11.84AWQLVIPTFKK184 pKa = 10.22TAVSYY189 pKa = 10.79SYY191 pKa = 11.38NDD193 pKa = 3.56GVVEE197 pKa = 3.96YY198 pKa = 10.58DD199 pKa = 3.79DD200 pKa = 5.64FGLGMMFVPSGLAYY214 pKa = 10.13FAGVNTGSAYY224 pKa = 10.83DD225 pKa = 3.52NLIFKK230 pKa = 10.62FEE232 pKa = 4.22LLQYY236 pKa = 10.36EE237 pKa = 4.77VVDD240 pKa = 4.35HH241 pKa = 7.41DD242 pKa = 4.9SDD244 pKa = 5.61GIPSWVEE251 pKa = 3.89DD252 pKa = 3.97LDD254 pKa = 5.86GDD256 pKa = 4.91LDD258 pKa = 4.15VSNDD262 pKa = 3.52DD263 pKa = 3.38TDD265 pKa = 5.06GNGFPNYY272 pKa = 9.79VDD274 pKa = 5.0RR275 pKa = 11.84DD276 pKa = 3.85DD277 pKa = 6.18DD278 pKa = 4.75GDD280 pKa = 4.05GVLTRR285 pKa = 11.84DD286 pKa = 3.45EE287 pKa = 5.71LIPTTYY293 pKa = 10.44TIDD296 pKa = 3.54LNAGEE301 pKa = 4.67EE302 pKa = 4.34EE303 pKa = 4.39PVLSEE308 pKa = 4.63GEE310 pKa = 4.3FEE312 pKa = 5.11VSRR315 pKa = 11.84DD316 pKa = 3.67LNNGILTIEE325 pKa = 4.37TVTIADD331 pKa = 3.96SNNDD335 pKa = 3.08GLADD339 pKa = 3.76YY340 pKa = 10.31LDD342 pKa = 4.17PNITINYY349 pKa = 9.07NEE351 pKa = 3.96
Molecular weight: 39.15 kDa
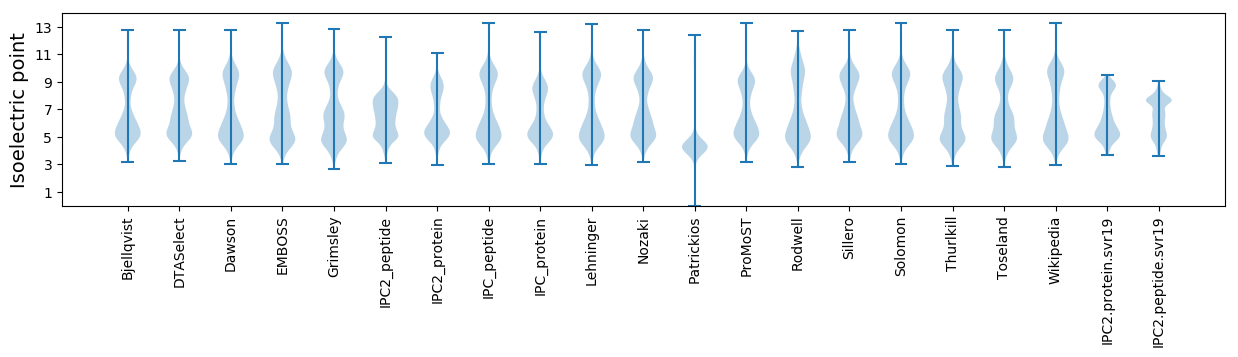
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2V4YH18|A0A2V4YH18_9FLAO 1-acyl-sn-glycerol-3-phosphate acyltransferase OS=Winogradskyella epiphytica OX=262005 GN=DFQ11_101645 PE=4 SV=1
MM1 pKa = 7.84PSGKK5 pKa = 9.32KK6 pKa = 9.59RR7 pKa = 11.84KK8 pKa = 7.05RR9 pKa = 11.84HH10 pKa = 5.11KK11 pKa = 10.59VATHH15 pKa = 5.21KK16 pKa = 10.3RR17 pKa = 11.84KK18 pKa = 9.62KK19 pKa = 9.04RR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84ANRR25 pKa = 11.84HH26 pKa = 4.5KK27 pKa = 10.78KK28 pKa = 9.37KK29 pKa = 10.83
MM1 pKa = 7.84PSGKK5 pKa = 9.32KK6 pKa = 9.59RR7 pKa = 11.84KK8 pKa = 7.05RR9 pKa = 11.84HH10 pKa = 5.11KK11 pKa = 10.59VATHH15 pKa = 5.21KK16 pKa = 10.3RR17 pKa = 11.84KK18 pKa = 9.62KK19 pKa = 9.04RR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84ANRR25 pKa = 11.84HH26 pKa = 4.5KK27 pKa = 10.78KK28 pKa = 9.37KK29 pKa = 10.83
Molecular weight: 3.63 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
994994 |
29 |
2881 |
330.9 |
37.38 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.247 ± 0.04 | 0.738 ± 0.019 |
5.837 ± 0.039 | 6.752 ± 0.041 |
5.141 ± 0.035 | 6.156 ± 0.041 |
1.831 ± 0.023 | 8.047 ± 0.045 |
7.701 ± 0.068 | 9.307 ± 0.059 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.204 ± 0.023 | 6.143 ± 0.048 |
3.305 ± 0.024 | 3.328 ± 0.025 |
3.439 ± 0.028 | 6.647 ± 0.044 |
5.882 ± 0.068 | 6.205 ± 0.036 |
0.978 ± 0.016 | 4.112 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
