
Phaeosphaeria nodorum (strain SN15 / ATCC MYA-4574 / FGSC 10173) (Glume blotch fungus) (Parastagonospora nodorum)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; dothideomyceta; Dothideomycetes; Pleosporomycetidae; Pleosporales; Pleosporineae; Phaeosphaeriaceae; Parastagonospora; Parastagonospora nodorum
Average proteome isoelectric point is 6.64
Get precalculated fractions of proteins
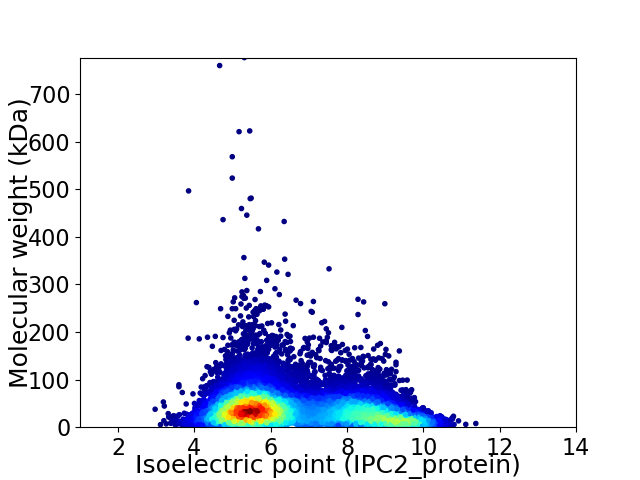
Virtual 2D-PAGE plot for 15998 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q0V2I3|Q0V2I3_PHANO Uncharacterized protein OS=Phaeosphaeria nodorum (strain SN15 / ATCC MYA-4574 / FGSC 10173) OX=321614 GN=SNOG_01781 PE=4 SV=2
MM1 pKa = 7.53AAATHH6 pKa = 6.65AVRR9 pKa = 11.84KK10 pKa = 9.83LDD12 pKa = 3.79AATDD16 pKa = 4.47DD17 pKa = 4.61IMDD20 pKa = 4.21LHH22 pKa = 7.55SDD24 pKa = 3.44SGLDD28 pKa = 4.33FDD30 pKa = 6.49DD31 pKa = 5.76GDD33 pKa = 4.23IDD35 pKa = 6.16LDD37 pKa = 4.48LDD39 pKa = 4.03PAPPAQADD47 pKa = 3.95DD48 pKa = 4.85DD49 pKa = 5.35DD50 pKa = 6.09ISLKK54 pKa = 10.88DD55 pKa = 3.54AGADD59 pKa = 3.33ADD61 pKa = 5.45LEE63 pKa = 4.57TQTVSPDD70 pKa = 2.91QDD72 pKa = 3.79MVDD75 pKa = 4.07HH76 pKa = 6.55EE77 pKa = 4.8VDD79 pKa = 4.09DD80 pKa = 5.03EE81 pKa = 4.85GDD83 pKa = 3.32IQLGDD88 pKa = 4.71GIHH91 pKa = 7.41DD92 pKa = 3.67DD93 pKa = 4.51HH94 pKa = 9.39VPIDD98 pKa = 4.0YY99 pKa = 7.7TTPAQEE105 pKa = 4.1TAPAHH110 pKa = 6.62PDD112 pKa = 3.48DD113 pKa = 5.88DD114 pKa = 6.36LIDD117 pKa = 3.99YY118 pKa = 11.09SDD120 pKa = 5.63DD121 pKa = 3.26EE122 pKa = 5.72DD123 pKa = 4.1IYY125 pKa = 10.34PTPTYY130 pKa = 10.71NQGTPAQSVGQTQDD144 pKa = 3.19VQNDD148 pKa = 3.29EE149 pKa = 4.5SASEE153 pKa = 3.96IHH155 pKa = 6.64ALPAEE160 pKa = 4.11AQIPGSTNEE169 pKa = 4.83DD170 pKa = 3.59EE171 pKa = 4.62IANHH175 pKa = 6.51GDD177 pKa = 3.56YY178 pKa = 11.39ADD180 pKa = 4.57GEE182 pKa = 5.13DD183 pKa = 3.67NFEE186 pKa = 4.12PSAEE190 pKa = 3.98NEE192 pKa = 4.22EE193 pKa = 4.11QHH195 pKa = 7.08DD196 pKa = 4.27EE197 pKa = 4.39NNFEE201 pKa = 4.3PSAEE205 pKa = 3.99NQEE208 pKa = 4.14QHH210 pKa = 7.28DD211 pKa = 4.08EE212 pKa = 4.52TEE214 pKa = 4.64FRR216 pKa = 11.84AQDD219 pKa = 3.34EE220 pKa = 4.74AEE222 pKa = 4.12YY223 pKa = 8.87DD224 pKa = 3.52TDD226 pKa = 4.17GDD228 pKa = 3.93TGGVPLQDD236 pKa = 3.45QDD238 pKa = 3.75SQADD242 pKa = 3.46VSEE245 pKa = 4.43YY246 pKa = 9.98QQGDD250 pKa = 3.67HH251 pKa = 6.93VEE253 pKa = 4.13QQSIEE258 pKa = 4.12SRR260 pKa = 11.84PITINYY266 pKa = 7.46EE267 pKa = 4.38GNEE270 pKa = 3.56LWLFKK275 pKa = 10.72QHH277 pKa = 7.24DD278 pKa = 4.08VDD280 pKa = 5.85EE281 pKa = 5.31SGDD284 pKa = 3.51WLTEE288 pKa = 3.84DD289 pKa = 3.55TSIIHH294 pKa = 6.35SSLSEE299 pKa = 3.85MFQACRR305 pKa = 11.84ASLGEE310 pKa = 4.23NISNEE315 pKa = 4.22TEE317 pKa = 3.52LGLRR321 pKa = 11.84FDD323 pKa = 4.7HH324 pKa = 6.14FHH326 pKa = 7.02NMEE329 pKa = 3.94IFEE332 pKa = 5.24DD333 pKa = 3.86STACVAVSLGRR344 pKa = 11.84MIDD347 pKa = 3.69LYY349 pKa = 10.25YY350 pKa = 10.4TLHH353 pKa = 6.08AQDD356 pKa = 4.94GEE358 pKa = 4.53SEE360 pKa = 4.3PEE362 pKa = 3.94SFYY365 pKa = 10.81MCLLSRR371 pKa = 11.84PRR373 pKa = 11.84FATLLSDD380 pKa = 3.7IAKK383 pKa = 9.87HH384 pKa = 6.25ADD386 pKa = 3.14QGHH389 pKa = 6.8GYY391 pKa = 10.13SGLNSAIAAGEE402 pKa = 4.13THH404 pKa = 6.73FVDD407 pKa = 3.4AHH409 pKa = 6.33SGQSTEE415 pKa = 4.43HH416 pKa = 7.01EE417 pKa = 4.39GTEE420 pKa = 3.84WDD422 pKa = 3.49QDD424 pKa = 3.59AEE426 pKa = 4.29GQEE429 pKa = 4.3EE430 pKa = 4.57YY431 pKa = 10.85GQKK434 pKa = 10.55EE435 pKa = 3.93EE436 pKa = 4.44TEE438 pKa = 4.07QEE440 pKa = 4.05EE441 pKa = 4.66NVAQEE446 pKa = 4.01NAQEE450 pKa = 4.12GTEE453 pKa = 4.24IVDD456 pKa = 3.67EE457 pKa = 4.73AADD460 pKa = 3.79EE461 pKa = 4.29AATGIDD467 pKa = 3.9LGEE470 pKa = 5.28NDD472 pKa = 5.18DD473 pKa = 4.31QDD475 pKa = 3.87NDD477 pKa = 3.49EE478 pKa = 4.35PRR480 pKa = 11.84VNGEE484 pKa = 3.85EE485 pKa = 4.12HH486 pKa = 6.52SSKK489 pKa = 11.32AEE491 pKa = 4.09TKK493 pKa = 10.08SEE495 pKa = 4.15HH496 pKa = 6.65ASTPAAVAHH505 pKa = 6.42GSSPAAQEE513 pKa = 4.21SPDD516 pKa = 3.45RR517 pKa = 11.84SAEE520 pKa = 4.19TTQHH524 pKa = 6.94DD525 pKa = 3.84VTANQTAEE533 pKa = 3.85QLANDD538 pKa = 4.05TVDD541 pKa = 3.51YY542 pKa = 11.37SDD544 pKa = 6.17DD545 pKa = 4.03EE546 pKa = 5.72DD547 pKa = 6.49DD548 pKa = 4.64EE549 pKa = 4.93PQTQAPTNASPSSTTVQGDD568 pKa = 3.46NPATIEE574 pKa = 3.98ASAPTAEE581 pKa = 4.2STSFEE586 pKa = 4.23KK587 pKa = 10.42EE588 pKa = 3.94YY589 pKa = 9.64QTGYY593 pKa = 11.43DD594 pKa = 3.45DD595 pKa = 4.87AFPYY599 pKa = 9.75AANGAYY605 pKa = 9.67EE606 pKa = 5.08DD607 pKa = 4.12YY608 pKa = 11.39AHH610 pKa = 7.07NEE612 pKa = 3.84EE613 pKa = 5.45QFDD616 pKa = 4.04HH617 pKa = 6.81EE618 pKa = 5.07NNADD622 pKa = 3.49MYY624 pKa = 10.96QEE626 pKa = 3.93YY627 pKa = 10.59DD628 pKa = 3.12GTYY631 pKa = 10.94GEE633 pKa = 5.48GEE635 pKa = 4.15ASDD638 pKa = 5.04LQTNDD643 pKa = 2.36QDD645 pKa = 4.3AYY647 pKa = 9.95PAEE650 pKa = 4.88ADD652 pKa = 4.31LDD654 pKa = 3.95GLTNQDD660 pKa = 3.31YY661 pKa = 9.87TGYY664 pKa = 10.67DD665 pKa = 3.65YY666 pKa = 11.52QDD668 pKa = 4.73LEE670 pKa = 4.18QQLQNEE676 pKa = 5.26FISGGEE682 pKa = 3.83VDD684 pKa = 3.58GTGAGEE690 pKa = 4.38SANGAHH696 pKa = 6.52EE697 pKa = 5.1LVDD700 pKa = 4.38GDD702 pKa = 5.62DD703 pKa = 5.43FLDD706 pKa = 4.71LEE708 pKa = 4.89NATEE712 pKa = 3.89WGTDD716 pKa = 2.85QGQAVEE722 pKa = 4.14VSGDD726 pKa = 3.69NIFLDD731 pKa = 4.45DD732 pKa = 4.56EE733 pKa = 4.7VTAHH737 pKa = 6.23YY738 pKa = 10.23EE739 pKa = 3.99EE740 pKa = 5.16EE741 pKa = 4.07EE742 pKa = 4.62GGVVEE747 pKa = 4.74QPAVAASSAADD758 pKa = 3.44PVAASSTDD766 pKa = 3.69LQDD769 pKa = 5.59SSPQGQKK776 pKa = 10.13RR777 pKa = 11.84SRR779 pKa = 11.84DD780 pKa = 3.52EE781 pKa = 4.94ASDD784 pKa = 3.94SVGDD788 pKa = 3.98ALDD791 pKa = 3.72PSGTHH796 pKa = 6.86HH797 pKa = 5.98IHH799 pKa = 7.49GINLCWQDD807 pKa = 3.2ADD809 pKa = 4.98FVFPDD814 pKa = 3.73TKK816 pKa = 10.46RR817 pKa = 11.84PRR819 pKa = 11.84VV820 pKa = 3.4
MM1 pKa = 7.53AAATHH6 pKa = 6.65AVRR9 pKa = 11.84KK10 pKa = 9.83LDD12 pKa = 3.79AATDD16 pKa = 4.47DD17 pKa = 4.61IMDD20 pKa = 4.21LHH22 pKa = 7.55SDD24 pKa = 3.44SGLDD28 pKa = 4.33FDD30 pKa = 6.49DD31 pKa = 5.76GDD33 pKa = 4.23IDD35 pKa = 6.16LDD37 pKa = 4.48LDD39 pKa = 4.03PAPPAQADD47 pKa = 3.95DD48 pKa = 4.85DD49 pKa = 5.35DD50 pKa = 6.09ISLKK54 pKa = 10.88DD55 pKa = 3.54AGADD59 pKa = 3.33ADD61 pKa = 5.45LEE63 pKa = 4.57TQTVSPDD70 pKa = 2.91QDD72 pKa = 3.79MVDD75 pKa = 4.07HH76 pKa = 6.55EE77 pKa = 4.8VDD79 pKa = 4.09DD80 pKa = 5.03EE81 pKa = 4.85GDD83 pKa = 3.32IQLGDD88 pKa = 4.71GIHH91 pKa = 7.41DD92 pKa = 3.67DD93 pKa = 4.51HH94 pKa = 9.39VPIDD98 pKa = 4.0YY99 pKa = 7.7TTPAQEE105 pKa = 4.1TAPAHH110 pKa = 6.62PDD112 pKa = 3.48DD113 pKa = 5.88DD114 pKa = 6.36LIDD117 pKa = 3.99YY118 pKa = 11.09SDD120 pKa = 5.63DD121 pKa = 3.26EE122 pKa = 5.72DD123 pKa = 4.1IYY125 pKa = 10.34PTPTYY130 pKa = 10.71NQGTPAQSVGQTQDD144 pKa = 3.19VQNDD148 pKa = 3.29EE149 pKa = 4.5SASEE153 pKa = 3.96IHH155 pKa = 6.64ALPAEE160 pKa = 4.11AQIPGSTNEE169 pKa = 4.83DD170 pKa = 3.59EE171 pKa = 4.62IANHH175 pKa = 6.51GDD177 pKa = 3.56YY178 pKa = 11.39ADD180 pKa = 4.57GEE182 pKa = 5.13DD183 pKa = 3.67NFEE186 pKa = 4.12PSAEE190 pKa = 3.98NEE192 pKa = 4.22EE193 pKa = 4.11QHH195 pKa = 7.08DD196 pKa = 4.27EE197 pKa = 4.39NNFEE201 pKa = 4.3PSAEE205 pKa = 3.99NQEE208 pKa = 4.14QHH210 pKa = 7.28DD211 pKa = 4.08EE212 pKa = 4.52TEE214 pKa = 4.64FRR216 pKa = 11.84AQDD219 pKa = 3.34EE220 pKa = 4.74AEE222 pKa = 4.12YY223 pKa = 8.87DD224 pKa = 3.52TDD226 pKa = 4.17GDD228 pKa = 3.93TGGVPLQDD236 pKa = 3.45QDD238 pKa = 3.75SQADD242 pKa = 3.46VSEE245 pKa = 4.43YY246 pKa = 9.98QQGDD250 pKa = 3.67HH251 pKa = 6.93VEE253 pKa = 4.13QQSIEE258 pKa = 4.12SRR260 pKa = 11.84PITINYY266 pKa = 7.46EE267 pKa = 4.38GNEE270 pKa = 3.56LWLFKK275 pKa = 10.72QHH277 pKa = 7.24DD278 pKa = 4.08VDD280 pKa = 5.85EE281 pKa = 5.31SGDD284 pKa = 3.51WLTEE288 pKa = 3.84DD289 pKa = 3.55TSIIHH294 pKa = 6.35SSLSEE299 pKa = 3.85MFQACRR305 pKa = 11.84ASLGEE310 pKa = 4.23NISNEE315 pKa = 4.22TEE317 pKa = 3.52LGLRR321 pKa = 11.84FDD323 pKa = 4.7HH324 pKa = 6.14FHH326 pKa = 7.02NMEE329 pKa = 3.94IFEE332 pKa = 5.24DD333 pKa = 3.86STACVAVSLGRR344 pKa = 11.84MIDD347 pKa = 3.69LYY349 pKa = 10.25YY350 pKa = 10.4TLHH353 pKa = 6.08AQDD356 pKa = 4.94GEE358 pKa = 4.53SEE360 pKa = 4.3PEE362 pKa = 3.94SFYY365 pKa = 10.81MCLLSRR371 pKa = 11.84PRR373 pKa = 11.84FATLLSDD380 pKa = 3.7IAKK383 pKa = 9.87HH384 pKa = 6.25ADD386 pKa = 3.14QGHH389 pKa = 6.8GYY391 pKa = 10.13SGLNSAIAAGEE402 pKa = 4.13THH404 pKa = 6.73FVDD407 pKa = 3.4AHH409 pKa = 6.33SGQSTEE415 pKa = 4.43HH416 pKa = 7.01EE417 pKa = 4.39GTEE420 pKa = 3.84WDD422 pKa = 3.49QDD424 pKa = 3.59AEE426 pKa = 4.29GQEE429 pKa = 4.3EE430 pKa = 4.57YY431 pKa = 10.85GQKK434 pKa = 10.55EE435 pKa = 3.93EE436 pKa = 4.44TEE438 pKa = 4.07QEE440 pKa = 4.05EE441 pKa = 4.66NVAQEE446 pKa = 4.01NAQEE450 pKa = 4.12GTEE453 pKa = 4.24IVDD456 pKa = 3.67EE457 pKa = 4.73AADD460 pKa = 3.79EE461 pKa = 4.29AATGIDD467 pKa = 3.9LGEE470 pKa = 5.28NDD472 pKa = 5.18DD473 pKa = 4.31QDD475 pKa = 3.87NDD477 pKa = 3.49EE478 pKa = 4.35PRR480 pKa = 11.84VNGEE484 pKa = 3.85EE485 pKa = 4.12HH486 pKa = 6.52SSKK489 pKa = 11.32AEE491 pKa = 4.09TKK493 pKa = 10.08SEE495 pKa = 4.15HH496 pKa = 6.65ASTPAAVAHH505 pKa = 6.42GSSPAAQEE513 pKa = 4.21SPDD516 pKa = 3.45RR517 pKa = 11.84SAEE520 pKa = 4.19TTQHH524 pKa = 6.94DD525 pKa = 3.84VTANQTAEE533 pKa = 3.85QLANDD538 pKa = 4.05TVDD541 pKa = 3.51YY542 pKa = 11.37SDD544 pKa = 6.17DD545 pKa = 4.03EE546 pKa = 5.72DD547 pKa = 6.49DD548 pKa = 4.64EE549 pKa = 4.93PQTQAPTNASPSSTTVQGDD568 pKa = 3.46NPATIEE574 pKa = 3.98ASAPTAEE581 pKa = 4.2STSFEE586 pKa = 4.23KK587 pKa = 10.42EE588 pKa = 3.94YY589 pKa = 9.64QTGYY593 pKa = 11.43DD594 pKa = 3.45DD595 pKa = 4.87AFPYY599 pKa = 9.75AANGAYY605 pKa = 9.67EE606 pKa = 5.08DD607 pKa = 4.12YY608 pKa = 11.39AHH610 pKa = 7.07NEE612 pKa = 3.84EE613 pKa = 5.45QFDD616 pKa = 4.04HH617 pKa = 6.81EE618 pKa = 5.07NNADD622 pKa = 3.49MYY624 pKa = 10.96QEE626 pKa = 3.93YY627 pKa = 10.59DD628 pKa = 3.12GTYY631 pKa = 10.94GEE633 pKa = 5.48GEE635 pKa = 4.15ASDD638 pKa = 5.04LQTNDD643 pKa = 2.36QDD645 pKa = 4.3AYY647 pKa = 9.95PAEE650 pKa = 4.88ADD652 pKa = 4.31LDD654 pKa = 3.95GLTNQDD660 pKa = 3.31YY661 pKa = 9.87TGYY664 pKa = 10.67DD665 pKa = 3.65YY666 pKa = 11.52QDD668 pKa = 4.73LEE670 pKa = 4.18QQLQNEE676 pKa = 5.26FISGGEE682 pKa = 3.83VDD684 pKa = 3.58GTGAGEE690 pKa = 4.38SANGAHH696 pKa = 6.52EE697 pKa = 5.1LVDD700 pKa = 4.38GDD702 pKa = 5.62DD703 pKa = 5.43FLDD706 pKa = 4.71LEE708 pKa = 4.89NATEE712 pKa = 3.89WGTDD716 pKa = 2.85QGQAVEE722 pKa = 4.14VSGDD726 pKa = 3.69NIFLDD731 pKa = 4.45DD732 pKa = 4.56EE733 pKa = 4.7VTAHH737 pKa = 6.23YY738 pKa = 10.23EE739 pKa = 3.99EE740 pKa = 5.16EE741 pKa = 4.07EE742 pKa = 4.62GGVVEE747 pKa = 4.74QPAVAASSAADD758 pKa = 3.44PVAASSTDD766 pKa = 3.69LQDD769 pKa = 5.59SSPQGQKK776 pKa = 10.13RR777 pKa = 11.84SRR779 pKa = 11.84DD780 pKa = 3.52EE781 pKa = 4.94ASDD784 pKa = 3.94SVGDD788 pKa = 3.98ALDD791 pKa = 3.72PSGTHH796 pKa = 6.86HH797 pKa = 5.98IHH799 pKa = 7.49GINLCWQDD807 pKa = 3.2ADD809 pKa = 4.98FVFPDD814 pKa = 3.73TKK816 pKa = 10.46RR817 pKa = 11.84PRR819 pKa = 11.84VV820 pKa = 3.4
Molecular weight: 89.17 kDa
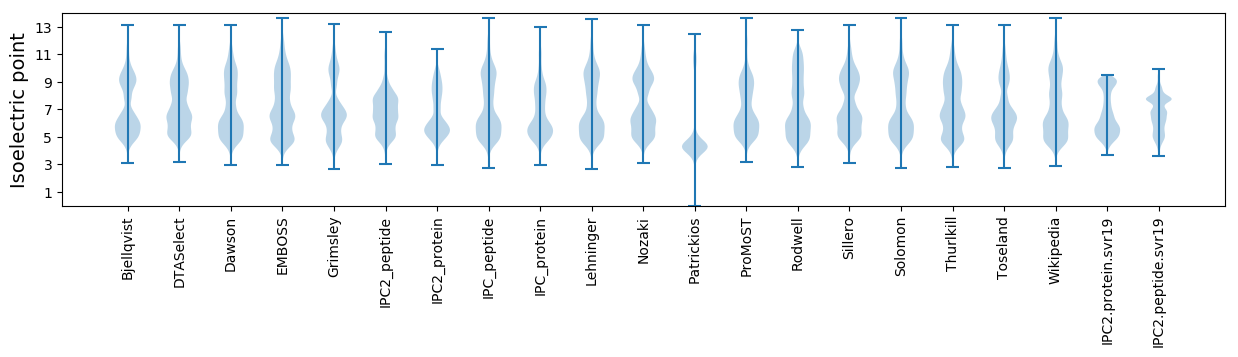
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q0UFA5|Q0UFA5_PHANO Uncharacterized protein OS=Phaeosphaeria nodorum (strain SN15 / ATCC MYA-4574 / FGSC 10173) OX=321614 GN=SNOG_09559 PE=4 SV=1
MM1 pKa = 7.37QIFGNSSRR9 pKa = 11.84GRR11 pKa = 11.84RR12 pKa = 11.84LRR14 pKa = 11.84WKK16 pKa = 10.5LRR18 pKa = 11.84LLTQSTTSRR27 pKa = 11.84ARR29 pKa = 11.84SRR31 pKa = 11.84TRR33 pKa = 11.84RR34 pKa = 11.84AFPRR38 pKa = 11.84TSSVSFSPASSSRR51 pKa = 11.84MAALFRR57 pKa = 11.84TTTSRR62 pKa = 11.84RR63 pKa = 11.84SPPSII68 pKa = 3.88
MM1 pKa = 7.37QIFGNSSRR9 pKa = 11.84GRR11 pKa = 11.84RR12 pKa = 11.84LRR14 pKa = 11.84WKK16 pKa = 10.5LRR18 pKa = 11.84LLTQSTTSRR27 pKa = 11.84ARR29 pKa = 11.84SRR31 pKa = 11.84TRR33 pKa = 11.84RR34 pKa = 11.84AFPRR38 pKa = 11.84TSSVSFSPASSSRR51 pKa = 11.84MAALFRR57 pKa = 11.84TTTSRR62 pKa = 11.84RR63 pKa = 11.84SPPSII68 pKa = 3.88
Molecular weight: 7.76 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
6077303 |
21 |
7045 |
379.9 |
42.08 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.89 ± 0.019 | 1.278 ± 0.007 |
5.729 ± 0.015 | 6.21 ± 0.019 |
3.706 ± 0.014 | 6.747 ± 0.02 |
2.468 ± 0.01 | 4.868 ± 0.015 |
5.131 ± 0.02 | 8.62 ± 0.022 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.333 ± 0.008 | 3.717 ± 0.013 |
5.93 ± 0.02 | 4.068 ± 0.015 |
5.97 ± 0.017 | 7.922 ± 0.024 |
6.07 ± 0.017 | 6.084 ± 0.014 |
1.495 ± 0.008 | 2.765 ± 0.01 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
