
Bos taurus papillomavirus 20
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; unclassified Papillomaviridae; non-primate mammal papillomaviruses
Average proteome isoelectric point is 6.54
Get precalculated fractions of proteins
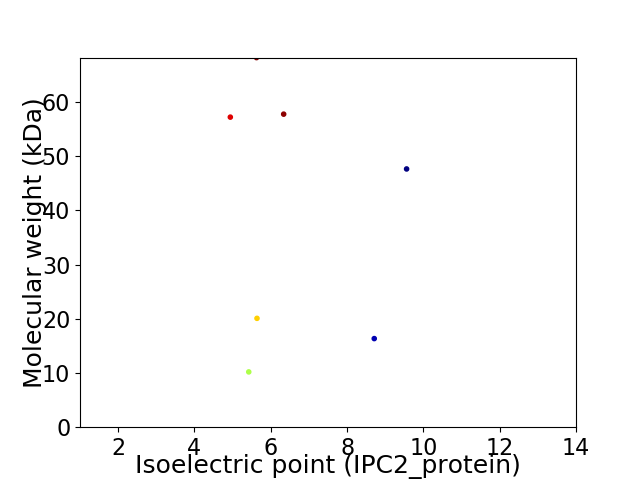
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1B2K242|A0A1B2K242_9PAPI E4 protein OS=Bos taurus papillomavirus 20 OX=1887218 GN=E4 PE=4 SV=1
MM1 pKa = 7.24VRR3 pKa = 11.84AQRR6 pKa = 11.84RR7 pKa = 11.84KK8 pKa = 9.5RR9 pKa = 11.84ASEE12 pKa = 3.67DD13 pKa = 2.98TLYY16 pKa = 10.56RR17 pKa = 11.84GCLAGQDD24 pKa = 4.1CPPDD28 pKa = 3.18IKK30 pKa = 10.94AKK32 pKa = 10.74YY33 pKa = 8.54EE34 pKa = 3.8QDD36 pKa = 3.29TLADD40 pKa = 5.53RR41 pKa = 11.84ILKK44 pKa = 8.66WVSTFLYY51 pKa = 9.77FGHH54 pKa = 7.29LGISSGKK61 pKa = 8.13GTGGQGGYY69 pKa = 8.03TRR71 pKa = 11.84LGGSGVSGGKK81 pKa = 7.96GTNVTRR87 pKa = 11.84PTVLVDD93 pKa = 3.71ALPPPGVPIDD103 pKa = 3.93TVSPDD108 pKa = 3.12SSIIPLLTDD117 pKa = 2.93SGGATTTDD125 pKa = 5.27LIPGTGEE132 pKa = 3.57IEE134 pKa = 4.2IVAEE138 pKa = 3.91VHH140 pKa = 6.31PPPTNGQDD148 pKa = 3.31SVIIGLEE155 pKa = 3.8PEE157 pKa = 4.29PPVLEE162 pKa = 4.21VTPEE166 pKa = 3.85HH167 pKa = 6.3TPIAPRR173 pKa = 11.84TRR175 pKa = 11.84TTVSKK180 pKa = 10.74HH181 pKa = 6.31DD182 pKa = 3.66NPAFNAYY189 pKa = 9.0VASTQLPAEE198 pKa = 4.29TAASDD203 pKa = 3.7NVHH206 pKa = 6.43IFHH209 pKa = 7.07GFGGEE214 pKa = 3.99IIGATEE220 pKa = 3.9GATFEE225 pKa = 4.66EE226 pKa = 5.01IPLEE230 pKa = 4.2EE231 pKa = 4.87FNPPTQGPSSSTPEE245 pKa = 3.71SGFRR249 pKa = 11.84GVLDD253 pKa = 3.74RR254 pKa = 11.84FQRR257 pKa = 11.84RR258 pKa = 11.84LYY260 pKa = 10.47NRR262 pKa = 11.84RR263 pKa = 11.84LVQQVKK269 pKa = 5.96VTRR272 pKa = 11.84RR273 pKa = 11.84SFLDD277 pKa = 3.5RR278 pKa = 11.84PSSLVRR284 pKa = 11.84WEE286 pKa = 4.35FDD288 pKa = 3.06NPTYY292 pKa = 10.95EE293 pKa = 5.21DD294 pKa = 4.16DD295 pKa = 3.77VSLLFEE301 pKa = 4.15QDD303 pKa = 3.3VHH305 pKa = 6.65NVSAAPDD312 pKa = 3.85LDD314 pKa = 3.79FQDD317 pKa = 3.75VVKK320 pKa = 10.78LSRR323 pKa = 11.84PHH325 pKa = 7.77IYY327 pKa = 10.15EE328 pKa = 3.66KK329 pKa = 10.56EE330 pKa = 4.15GYY332 pKa = 9.3IRR334 pKa = 11.84LSRR337 pKa = 11.84LGKK340 pKa = 9.98KK341 pKa = 8.77ATISTRR347 pKa = 11.84SGTTIGSQVHH357 pKa = 5.95YY358 pKa = 10.39YY359 pKa = 10.89SDD361 pKa = 4.34LSPIQPVEE369 pKa = 4.44EE370 pKa = 4.46IPLHH374 pKa = 5.56TFSSTDD380 pKa = 2.84RR381 pKa = 11.84SVIMQPLEE389 pKa = 3.63EE390 pKa = 4.15SVIIGSEE397 pKa = 4.11YY398 pKa = 10.06FDD400 pKa = 4.22EE401 pKa = 4.54NAVIYY406 pKa = 11.1NEE408 pKa = 4.23DD409 pKa = 3.51TSLLPEE415 pKa = 4.4FADD418 pKa = 5.21DD419 pKa = 4.94VLDD422 pKa = 4.96DD423 pKa = 4.63PYY425 pKa = 11.6EE426 pKa = 4.09EE427 pKa = 5.68DD428 pKa = 3.6FANTRR433 pKa = 11.84LEE435 pKa = 4.1VSGSSSRR442 pKa = 11.84QTIYY446 pKa = 10.97VPDD449 pKa = 5.59GIPPGSVRR457 pKa = 11.84AFVTDD462 pKa = 3.73YY463 pKa = 11.05NNHH466 pKa = 5.13STVTPEE472 pKa = 4.33GSNPLQPDD480 pKa = 4.07IIHH483 pKa = 7.2PDD485 pKa = 3.16IVPDD489 pKa = 4.65IILDD493 pKa = 3.75IYY495 pKa = 10.35TPGSTFYY502 pKa = 10.65LHH504 pKa = 6.96PSHH507 pKa = 6.28YY508 pKa = 9.65RR509 pKa = 11.84RR510 pKa = 11.84RR511 pKa = 11.84GRR513 pKa = 11.84KK514 pKa = 8.75RR515 pKa = 11.84KK516 pKa = 8.64RR517 pKa = 11.84TVFF520 pKa = 3.39
MM1 pKa = 7.24VRR3 pKa = 11.84AQRR6 pKa = 11.84RR7 pKa = 11.84KK8 pKa = 9.5RR9 pKa = 11.84ASEE12 pKa = 3.67DD13 pKa = 2.98TLYY16 pKa = 10.56RR17 pKa = 11.84GCLAGQDD24 pKa = 4.1CPPDD28 pKa = 3.18IKK30 pKa = 10.94AKK32 pKa = 10.74YY33 pKa = 8.54EE34 pKa = 3.8QDD36 pKa = 3.29TLADD40 pKa = 5.53RR41 pKa = 11.84ILKK44 pKa = 8.66WVSTFLYY51 pKa = 9.77FGHH54 pKa = 7.29LGISSGKK61 pKa = 8.13GTGGQGGYY69 pKa = 8.03TRR71 pKa = 11.84LGGSGVSGGKK81 pKa = 7.96GTNVTRR87 pKa = 11.84PTVLVDD93 pKa = 3.71ALPPPGVPIDD103 pKa = 3.93TVSPDD108 pKa = 3.12SSIIPLLTDD117 pKa = 2.93SGGATTTDD125 pKa = 5.27LIPGTGEE132 pKa = 3.57IEE134 pKa = 4.2IVAEE138 pKa = 3.91VHH140 pKa = 6.31PPPTNGQDD148 pKa = 3.31SVIIGLEE155 pKa = 3.8PEE157 pKa = 4.29PPVLEE162 pKa = 4.21VTPEE166 pKa = 3.85HH167 pKa = 6.3TPIAPRR173 pKa = 11.84TRR175 pKa = 11.84TTVSKK180 pKa = 10.74HH181 pKa = 6.31DD182 pKa = 3.66NPAFNAYY189 pKa = 9.0VASTQLPAEE198 pKa = 4.29TAASDD203 pKa = 3.7NVHH206 pKa = 6.43IFHH209 pKa = 7.07GFGGEE214 pKa = 3.99IIGATEE220 pKa = 3.9GATFEE225 pKa = 4.66EE226 pKa = 5.01IPLEE230 pKa = 4.2EE231 pKa = 4.87FNPPTQGPSSSTPEE245 pKa = 3.71SGFRR249 pKa = 11.84GVLDD253 pKa = 3.74RR254 pKa = 11.84FQRR257 pKa = 11.84RR258 pKa = 11.84LYY260 pKa = 10.47NRR262 pKa = 11.84RR263 pKa = 11.84LVQQVKK269 pKa = 5.96VTRR272 pKa = 11.84RR273 pKa = 11.84SFLDD277 pKa = 3.5RR278 pKa = 11.84PSSLVRR284 pKa = 11.84WEE286 pKa = 4.35FDD288 pKa = 3.06NPTYY292 pKa = 10.95EE293 pKa = 5.21DD294 pKa = 4.16DD295 pKa = 3.77VSLLFEE301 pKa = 4.15QDD303 pKa = 3.3VHH305 pKa = 6.65NVSAAPDD312 pKa = 3.85LDD314 pKa = 3.79FQDD317 pKa = 3.75VVKK320 pKa = 10.78LSRR323 pKa = 11.84PHH325 pKa = 7.77IYY327 pKa = 10.15EE328 pKa = 3.66KK329 pKa = 10.56EE330 pKa = 4.15GYY332 pKa = 9.3IRR334 pKa = 11.84LSRR337 pKa = 11.84LGKK340 pKa = 9.98KK341 pKa = 8.77ATISTRR347 pKa = 11.84SGTTIGSQVHH357 pKa = 5.95YY358 pKa = 10.39YY359 pKa = 10.89SDD361 pKa = 4.34LSPIQPVEE369 pKa = 4.44EE370 pKa = 4.46IPLHH374 pKa = 5.56TFSSTDD380 pKa = 2.84RR381 pKa = 11.84SVIMQPLEE389 pKa = 3.63EE390 pKa = 4.15SVIIGSEE397 pKa = 4.11YY398 pKa = 10.06FDD400 pKa = 4.22EE401 pKa = 4.54NAVIYY406 pKa = 11.1NEE408 pKa = 4.23DD409 pKa = 3.51TSLLPEE415 pKa = 4.4FADD418 pKa = 5.21DD419 pKa = 4.94VLDD422 pKa = 4.96DD423 pKa = 4.63PYY425 pKa = 11.6EE426 pKa = 4.09EE427 pKa = 5.68DD428 pKa = 3.6FANTRR433 pKa = 11.84LEE435 pKa = 4.1VSGSSSRR442 pKa = 11.84QTIYY446 pKa = 10.97VPDD449 pKa = 5.59GIPPGSVRR457 pKa = 11.84AFVTDD462 pKa = 3.73YY463 pKa = 11.05NNHH466 pKa = 5.13STVTPEE472 pKa = 4.33GSNPLQPDD480 pKa = 4.07IIHH483 pKa = 7.2PDD485 pKa = 3.16IVPDD489 pKa = 4.65IILDD493 pKa = 3.75IYY495 pKa = 10.35TPGSTFYY502 pKa = 10.65LHH504 pKa = 6.96PSHH507 pKa = 6.28YY508 pKa = 9.65RR509 pKa = 11.84RR510 pKa = 11.84RR511 pKa = 11.84GRR513 pKa = 11.84KK514 pKa = 8.75RR515 pKa = 11.84KK516 pKa = 8.64RR517 pKa = 11.84TVFF520 pKa = 3.39
Molecular weight: 57.2 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1B2K224|A0A1B2K224_9PAPI Replication protein E1 OS=Bos taurus papillomavirus 20 OX=1887218 GN=E1 PE=3 SV=1
MM1 pKa = 7.66EE2 pKa = 4.54EE3 pKa = 3.23LAARR7 pKa = 11.84FDD9 pKa = 4.08AVQEE13 pKa = 3.96RR14 pKa = 11.84LMQIYY19 pKa = 10.16EE20 pKa = 4.32SGSVTLEE27 pKa = 3.75SQIEE31 pKa = 4.34YY32 pKa = 6.25WTVVRR37 pKa = 11.84KK38 pKa = 9.83EE39 pKa = 3.55YY40 pKa = 11.08ALYY43 pKa = 8.85YY44 pKa = 8.01TARR47 pKa = 11.84KK48 pKa = 9.46QGIKK52 pKa = 10.45KK53 pKa = 10.04LGLYY57 pKa = 8.15VVPSLQIAEE66 pKa = 4.36QKK68 pKa = 10.58AKK70 pKa = 10.36EE71 pKa = 4.3AIRR74 pKa = 11.84MTLYY78 pKa = 10.94LEE80 pKa = 4.52SLSKK84 pKa = 10.96SQFGDD89 pKa = 3.8LQWTLPEE96 pKa = 4.14TSLEE100 pKa = 4.3TFSAPPANTFKK111 pKa = 11.1KK112 pKa = 10.13KK113 pKa = 9.63GAQAIVTYY121 pKa = 10.98DD122 pKa = 3.5NDD124 pKa = 3.83PEE126 pKa = 3.98NRR128 pKa = 11.84MMYY131 pKa = 8.24TVWKK135 pKa = 9.19EE136 pKa = 3.56LYY138 pKa = 9.37YY139 pKa = 10.47QDD141 pKa = 5.65DD142 pKa = 3.72EE143 pKa = 4.81EE144 pKa = 4.35MWHH147 pKa = 6.65KK148 pKa = 11.18GEE150 pKa = 4.15STVNHH155 pKa = 6.08EE156 pKa = 4.34GIYY159 pKa = 10.64YY160 pKa = 10.07VDD162 pKa = 3.14NTGRR166 pKa = 11.84TVYY169 pKa = 9.71YY170 pKa = 10.59QRR172 pKa = 11.84FAQDD176 pKa = 3.15AEE178 pKa = 4.38RR179 pKa = 11.84FSKK182 pKa = 10.28SGQWNVQYY190 pKa = 10.72EE191 pKa = 4.4NQVFSAPVTSSSRR204 pKa = 11.84QLRR207 pKa = 11.84TPRR210 pKa = 11.84GQGPEE215 pKa = 3.94EE216 pKa = 4.24RR217 pKa = 11.84QDD219 pKa = 3.24STGAQTHH226 pKa = 5.97SPAAGPRR233 pKa = 11.84QPAAPASRR241 pKa = 11.84RR242 pKa = 11.84PPVRR246 pKa = 11.84PSLRR250 pKa = 11.84GSRR253 pKa = 11.84GSLSSAEE260 pKa = 3.88RR261 pKa = 11.84GPRR264 pKa = 11.84RR265 pKa = 11.84GRR267 pKa = 11.84RR268 pKa = 11.84GPRR271 pKa = 11.84AGRR274 pKa = 11.84QTSTSDD280 pKa = 2.84GSSRR284 pKa = 11.84SRR286 pKa = 11.84SRR288 pKa = 11.84SRR290 pKa = 11.84SRR292 pKa = 11.84TRR294 pKa = 11.84SRR296 pKa = 11.84SRR298 pKa = 11.84SRR300 pKa = 11.84GPEE303 pKa = 3.69SGGQAPVSPEE313 pKa = 3.74EE314 pKa = 3.86VGTRR318 pKa = 11.84TRR320 pKa = 11.84TPEE323 pKa = 3.64TRR325 pKa = 11.84PTSRR329 pKa = 11.84IAQLIEE335 pKa = 4.05DD336 pKa = 4.11ARR338 pKa = 11.84DD339 pKa = 3.58PPVLLLQGPANSLKK353 pKa = 10.35CFRR356 pKa = 11.84RR357 pKa = 11.84RR358 pKa = 11.84SKK360 pKa = 8.73TQHH363 pKa = 6.53PSLFLCMSTSWSWVCRR379 pKa = 11.84TSTQKK384 pKa = 10.81LGNRR388 pKa = 11.84MLVAFSDD395 pKa = 3.9ASQRR399 pKa = 11.84TAFLSRR405 pKa = 11.84VKK407 pKa = 10.53LPRR410 pKa = 11.84GVTWTTGSLNGLL422 pKa = 3.82
MM1 pKa = 7.66EE2 pKa = 4.54EE3 pKa = 3.23LAARR7 pKa = 11.84FDD9 pKa = 4.08AVQEE13 pKa = 3.96RR14 pKa = 11.84LMQIYY19 pKa = 10.16EE20 pKa = 4.32SGSVTLEE27 pKa = 3.75SQIEE31 pKa = 4.34YY32 pKa = 6.25WTVVRR37 pKa = 11.84KK38 pKa = 9.83EE39 pKa = 3.55YY40 pKa = 11.08ALYY43 pKa = 8.85YY44 pKa = 8.01TARR47 pKa = 11.84KK48 pKa = 9.46QGIKK52 pKa = 10.45KK53 pKa = 10.04LGLYY57 pKa = 8.15VVPSLQIAEE66 pKa = 4.36QKK68 pKa = 10.58AKK70 pKa = 10.36EE71 pKa = 4.3AIRR74 pKa = 11.84MTLYY78 pKa = 10.94LEE80 pKa = 4.52SLSKK84 pKa = 10.96SQFGDD89 pKa = 3.8LQWTLPEE96 pKa = 4.14TSLEE100 pKa = 4.3TFSAPPANTFKK111 pKa = 11.1KK112 pKa = 10.13KK113 pKa = 9.63GAQAIVTYY121 pKa = 10.98DD122 pKa = 3.5NDD124 pKa = 3.83PEE126 pKa = 3.98NRR128 pKa = 11.84MMYY131 pKa = 8.24TVWKK135 pKa = 9.19EE136 pKa = 3.56LYY138 pKa = 9.37YY139 pKa = 10.47QDD141 pKa = 5.65DD142 pKa = 3.72EE143 pKa = 4.81EE144 pKa = 4.35MWHH147 pKa = 6.65KK148 pKa = 11.18GEE150 pKa = 4.15STVNHH155 pKa = 6.08EE156 pKa = 4.34GIYY159 pKa = 10.64YY160 pKa = 10.07VDD162 pKa = 3.14NTGRR166 pKa = 11.84TVYY169 pKa = 9.71YY170 pKa = 10.59QRR172 pKa = 11.84FAQDD176 pKa = 3.15AEE178 pKa = 4.38RR179 pKa = 11.84FSKK182 pKa = 10.28SGQWNVQYY190 pKa = 10.72EE191 pKa = 4.4NQVFSAPVTSSSRR204 pKa = 11.84QLRR207 pKa = 11.84TPRR210 pKa = 11.84GQGPEE215 pKa = 3.94EE216 pKa = 4.24RR217 pKa = 11.84QDD219 pKa = 3.24STGAQTHH226 pKa = 5.97SPAAGPRR233 pKa = 11.84QPAAPASRR241 pKa = 11.84RR242 pKa = 11.84PPVRR246 pKa = 11.84PSLRR250 pKa = 11.84GSRR253 pKa = 11.84GSLSSAEE260 pKa = 3.88RR261 pKa = 11.84GPRR264 pKa = 11.84RR265 pKa = 11.84GRR267 pKa = 11.84RR268 pKa = 11.84GPRR271 pKa = 11.84AGRR274 pKa = 11.84QTSTSDD280 pKa = 2.84GSSRR284 pKa = 11.84SRR286 pKa = 11.84SRR288 pKa = 11.84SRR290 pKa = 11.84SRR292 pKa = 11.84TRR294 pKa = 11.84SRR296 pKa = 11.84SRR298 pKa = 11.84SRR300 pKa = 11.84GPEE303 pKa = 3.69SGGQAPVSPEE313 pKa = 3.74EE314 pKa = 3.86VGTRR318 pKa = 11.84TRR320 pKa = 11.84TPEE323 pKa = 3.64TRR325 pKa = 11.84PTSRR329 pKa = 11.84IAQLIEE335 pKa = 4.05DD336 pKa = 4.11ARR338 pKa = 11.84DD339 pKa = 3.58PPVLLLQGPANSLKK353 pKa = 10.35CFRR356 pKa = 11.84RR357 pKa = 11.84RR358 pKa = 11.84SKK360 pKa = 8.73TQHH363 pKa = 6.53PSLFLCMSTSWSWVCRR379 pKa = 11.84TSTQKK384 pKa = 10.81LGNRR388 pKa = 11.84MLVAFSDD395 pKa = 3.9ASQRR399 pKa = 11.84TAFLSRR405 pKa = 11.84VKK407 pKa = 10.53LPRR410 pKa = 11.84GVTWTTGSLNGLL422 pKa = 3.82
Molecular weight: 47.64 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2458 |
93 |
598 |
351.1 |
39.63 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.021 ± 0.647 | 2.034 ± 0.603 |
5.858 ± 0.669 | 6.347 ± 0.3 |
4.434 ± 0.706 | 6.387 ± 0.724 |
1.993 ± 0.217 | 4.231 ± 0.613 |
5.614 ± 0.973 | 8.747 ± 0.74 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.749 ± 0.358 | 3.662 ± 0.495 |
6.876 ± 0.916 | 5.004 ± 0.401 |
6.509 ± 1.066 | 7.608 ± 0.805 |
6.021 ± 0.915 | 6.021 ± 0.418 |
1.424 ± 0.284 | 3.458 ± 0.208 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
