
Wenling frogfish filovirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Filoviridae; Striavirus; Xilang striavirus
Average proteome isoelectric point is 6.96
Get precalculated fractions of proteins
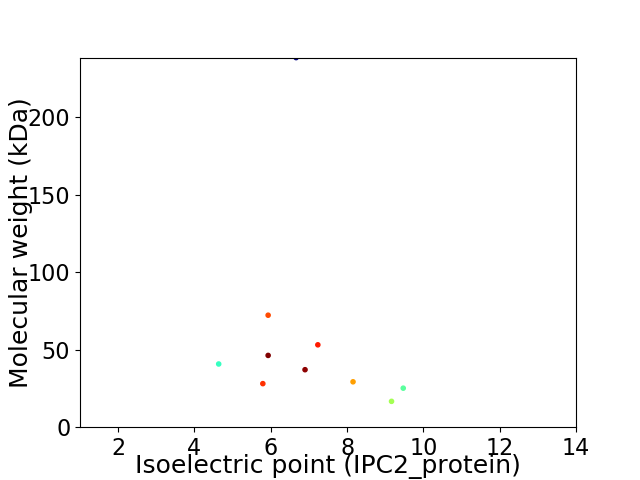
Virtual 2D-PAGE plot for 10 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2P1GML8|A0A2P1GML8_9MONO Nucleoprotein OS=Wenling frogfish filovirus OX=2116487 PE=3 SV=1
MM1 pKa = 8.07DD2 pKa = 5.25MDD4 pKa = 4.91TMDD7 pKa = 4.9LSCIAGSNTGGEE19 pKa = 4.26NPFQRR24 pKa = 11.84LDD26 pKa = 3.7GLPALQLGTISRR38 pKa = 11.84VRR40 pKa = 11.84DD41 pKa = 3.47LEE43 pKa = 4.08ATMEE47 pKa = 4.31TPQTQPQPQATTRR60 pKa = 11.84QNLADD65 pKa = 4.34DD66 pKa = 4.89VYY68 pKa = 10.87AQLGAEE74 pKa = 4.39TKK76 pKa = 10.52EE77 pKa = 4.37EE78 pKa = 4.47PDD80 pKa = 3.38DD81 pKa = 3.87QYY83 pKa = 11.96EE84 pKa = 4.05GDD86 pKa = 4.04EE87 pKa = 4.31PDD89 pKa = 4.95DD90 pKa = 3.68YY91 pKa = 11.5FSAIEE96 pKa = 4.34TIVGSCMKK104 pKa = 10.69GFAEE108 pKa = 4.67QLNQQGQHH116 pKa = 6.76LMILQKK122 pKa = 10.94KK123 pKa = 9.43LDD125 pKa = 3.87LVQVGVAEE133 pKa = 4.52IEE135 pKa = 3.98QSLRR139 pKa = 11.84ASTVTLADD147 pKa = 3.61EE148 pKa = 5.43LKK150 pKa = 9.87TDD152 pKa = 3.45MAVLVKK158 pKa = 10.05TISQVSAEE166 pKa = 4.53LKK168 pKa = 10.54NLVTITGPAAGTADD182 pKa = 3.16AVVRR186 pKa = 11.84CLSQGRR192 pKa = 11.84QLPDD196 pKa = 3.36VPCVISEE203 pKa = 4.3SSVPTIQHH211 pKa = 6.46LVATAPPTRR220 pKa = 11.84SAVDD224 pKa = 3.72LGAALDD230 pKa = 3.79MYY232 pKa = 9.83QTVEE236 pKa = 4.13EE237 pKa = 4.23EE238 pKa = 3.74QGEE241 pKa = 4.49NFGRR245 pKa = 11.84PVYY248 pKa = 10.59EE249 pKa = 4.06GDD251 pKa = 3.57EE252 pKa = 4.16LAEE255 pKa = 5.33DD256 pKa = 3.66ICSLITTRR264 pKa = 11.84NPALFKK270 pKa = 10.54KK271 pKa = 10.6IKK273 pKa = 10.56AEE275 pKa = 3.45VDD277 pKa = 3.09AAGEE281 pKa = 4.18EE282 pKa = 4.28QASRR286 pKa = 11.84IYY288 pKa = 10.75TEE290 pKa = 5.08LLPMDD295 pKa = 3.77NTSRR299 pKa = 11.84EE300 pKa = 3.88AYY302 pKa = 8.69IFQWAKK308 pKa = 8.8IQRR311 pKa = 11.84TEE313 pKa = 3.92KK314 pKa = 10.35HH315 pKa = 6.26HH316 pKa = 6.63LPHH319 pKa = 6.97PASGNHH325 pKa = 4.6QVTKK329 pKa = 10.39KK330 pKa = 9.7VVVMKK335 pKa = 8.84TPPGPEE341 pKa = 3.6YY342 pKa = 10.59MLMNRR347 pKa = 11.84KK348 pKa = 6.75PTKK351 pKa = 9.91RR352 pKa = 11.84DD353 pKa = 3.09LQEE356 pKa = 3.34FDD358 pKa = 5.36KK359 pKa = 10.85IVKK362 pKa = 10.14YY363 pKa = 10.58DD364 pKa = 3.4GQYY367 pKa = 11.08FSTT370 pKa = 4.36
MM1 pKa = 8.07DD2 pKa = 5.25MDD4 pKa = 4.91TMDD7 pKa = 4.9LSCIAGSNTGGEE19 pKa = 4.26NPFQRR24 pKa = 11.84LDD26 pKa = 3.7GLPALQLGTISRR38 pKa = 11.84VRR40 pKa = 11.84DD41 pKa = 3.47LEE43 pKa = 4.08ATMEE47 pKa = 4.31TPQTQPQPQATTRR60 pKa = 11.84QNLADD65 pKa = 4.34DD66 pKa = 4.89VYY68 pKa = 10.87AQLGAEE74 pKa = 4.39TKK76 pKa = 10.52EE77 pKa = 4.37EE78 pKa = 4.47PDD80 pKa = 3.38DD81 pKa = 3.87QYY83 pKa = 11.96EE84 pKa = 4.05GDD86 pKa = 4.04EE87 pKa = 4.31PDD89 pKa = 4.95DD90 pKa = 3.68YY91 pKa = 11.5FSAIEE96 pKa = 4.34TIVGSCMKK104 pKa = 10.69GFAEE108 pKa = 4.67QLNQQGQHH116 pKa = 6.76LMILQKK122 pKa = 10.94KK123 pKa = 9.43LDD125 pKa = 3.87LVQVGVAEE133 pKa = 4.52IEE135 pKa = 3.98QSLRR139 pKa = 11.84ASTVTLADD147 pKa = 3.61EE148 pKa = 5.43LKK150 pKa = 9.87TDD152 pKa = 3.45MAVLVKK158 pKa = 10.05TISQVSAEE166 pKa = 4.53LKK168 pKa = 10.54NLVTITGPAAGTADD182 pKa = 3.16AVVRR186 pKa = 11.84CLSQGRR192 pKa = 11.84QLPDD196 pKa = 3.36VPCVISEE203 pKa = 4.3SSVPTIQHH211 pKa = 6.46LVATAPPTRR220 pKa = 11.84SAVDD224 pKa = 3.72LGAALDD230 pKa = 3.79MYY232 pKa = 9.83QTVEE236 pKa = 4.13EE237 pKa = 4.23EE238 pKa = 3.74QGEE241 pKa = 4.49NFGRR245 pKa = 11.84PVYY248 pKa = 10.59EE249 pKa = 4.06GDD251 pKa = 3.57EE252 pKa = 4.16LAEE255 pKa = 5.33DD256 pKa = 3.66ICSLITTRR264 pKa = 11.84NPALFKK270 pKa = 10.54KK271 pKa = 10.6IKK273 pKa = 10.56AEE275 pKa = 3.45VDD277 pKa = 3.09AAGEE281 pKa = 4.18EE282 pKa = 4.28QASRR286 pKa = 11.84IYY288 pKa = 10.75TEE290 pKa = 5.08LLPMDD295 pKa = 3.77NTSRR299 pKa = 11.84EE300 pKa = 3.88AYY302 pKa = 8.69IFQWAKK308 pKa = 8.8IQRR311 pKa = 11.84TEE313 pKa = 3.92KK314 pKa = 10.35HH315 pKa = 6.26HH316 pKa = 6.63LPHH319 pKa = 6.97PASGNHH325 pKa = 4.6QVTKK329 pKa = 10.39KK330 pKa = 9.7VVVMKK335 pKa = 8.84TPPGPEE341 pKa = 3.6YY342 pKa = 10.59MLMNRR347 pKa = 11.84KK348 pKa = 6.75PTKK351 pKa = 9.91RR352 pKa = 11.84DD353 pKa = 3.09LQEE356 pKa = 3.34FDD358 pKa = 5.36KK359 pKa = 10.85IVKK362 pKa = 10.14YY363 pKa = 10.58DD364 pKa = 3.4GQYY367 pKa = 11.08FSTT370 pKa = 4.36
Molecular weight: 40.77 kDa
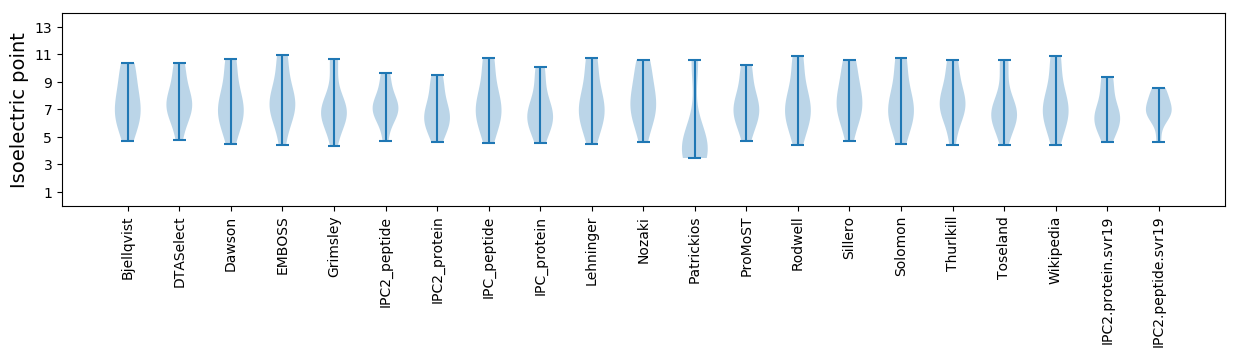
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2P1GML3|A0A2P1GML3_9MONO Putative glycoprotein 2 OS=Wenling frogfish filovirus OX=2116487 PE=4 SV=1
MM1 pKa = 7.01TASNPAAIALICMGCFLGLVLTWGLADD28 pKa = 5.31LICRR32 pKa = 11.84CKK34 pKa = 10.51RR35 pKa = 11.84RR36 pKa = 11.84AQKK39 pKa = 10.32QKK41 pKa = 10.68ARR43 pKa = 11.84RR44 pKa = 11.84ALPRR48 pKa = 11.84LRR50 pKa = 11.84IVSDD54 pKa = 3.83SSALTLQTDD63 pKa = 3.83VTSGLSSPNGSRR75 pKa = 11.84PISRR79 pKa = 11.84FSDD82 pKa = 4.0CNLHH86 pKa = 6.99HH87 pKa = 7.21PWAQEE92 pKa = 3.75KK93 pKa = 10.31LLHH96 pKa = 5.72IVLRR100 pKa = 11.84EE101 pKa = 3.94TTPLGDD107 pKa = 3.19IKK109 pKa = 10.86LVPFLRR115 pKa = 11.84LSHH118 pKa = 6.24HH119 pKa = 5.93QFSEE123 pKa = 3.74DD124 pKa = 3.33MKK126 pKa = 11.0IMRR129 pKa = 11.84SNLYY133 pKa = 10.74GFVGWLRR140 pKa = 11.84AEE142 pKa = 4.0CLIRR146 pKa = 11.84RR147 pKa = 11.84GSQQHH152 pKa = 5.6PVWSVVQTLNSPDD165 pKa = 3.66HH166 pKa = 6.42RR167 pKa = 11.84WAGSRR172 pKa = 11.84LKK174 pKa = 11.34GEE176 pKa = 4.38GFPLGKK182 pKa = 9.67ISQAAVLAVLDD193 pKa = 4.2VVVHH197 pKa = 6.49GSHH200 pKa = 6.64PHH202 pKa = 5.72LGYY205 pKa = 11.19ALRR208 pKa = 11.84HH209 pKa = 6.26HH210 pKa = 6.86IADD213 pKa = 4.16TYY215 pKa = 10.93RR216 pKa = 11.84PGVRR220 pKa = 11.84PRR222 pKa = 11.84TALII226 pKa = 3.81
MM1 pKa = 7.01TASNPAAIALICMGCFLGLVLTWGLADD28 pKa = 5.31LICRR32 pKa = 11.84CKK34 pKa = 10.51RR35 pKa = 11.84RR36 pKa = 11.84AQKK39 pKa = 10.32QKK41 pKa = 10.68ARR43 pKa = 11.84RR44 pKa = 11.84ALPRR48 pKa = 11.84LRR50 pKa = 11.84IVSDD54 pKa = 3.83SSALTLQTDD63 pKa = 3.83VTSGLSSPNGSRR75 pKa = 11.84PISRR79 pKa = 11.84FSDD82 pKa = 4.0CNLHH86 pKa = 6.99HH87 pKa = 7.21PWAQEE92 pKa = 3.75KK93 pKa = 10.31LLHH96 pKa = 5.72IVLRR100 pKa = 11.84EE101 pKa = 3.94TTPLGDD107 pKa = 3.19IKK109 pKa = 10.86LVPFLRR115 pKa = 11.84LSHH118 pKa = 6.24HH119 pKa = 5.93QFSEE123 pKa = 3.74DD124 pKa = 3.33MKK126 pKa = 11.0IMRR129 pKa = 11.84SNLYY133 pKa = 10.74GFVGWLRR140 pKa = 11.84AEE142 pKa = 4.0CLIRR146 pKa = 11.84RR147 pKa = 11.84GSQQHH152 pKa = 5.6PVWSVVQTLNSPDD165 pKa = 3.66HH166 pKa = 6.42RR167 pKa = 11.84WAGSRR172 pKa = 11.84LKK174 pKa = 11.34GEE176 pKa = 4.38GFPLGKK182 pKa = 9.67ISQAAVLAVLDD193 pKa = 4.2VVVHH197 pKa = 6.49GSHH200 pKa = 6.64PHH202 pKa = 5.72LGYY205 pKa = 11.19ALRR208 pKa = 11.84HH209 pKa = 6.26HH210 pKa = 6.86IADD213 pKa = 4.16TYY215 pKa = 10.93RR216 pKa = 11.84PGVRR220 pKa = 11.84PRR222 pKa = 11.84TALII226 pKa = 3.81
Molecular weight: 25.18 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5282 |
151 |
2116 |
528.2 |
58.72 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.819 ± 0.332 | 2.007 ± 0.22 |
5.168 ± 0.409 | 5.566 ± 0.437 |
3.597 ± 0.429 | 6.285 ± 0.352 |
2.897 ± 0.315 | 4.979 ± 0.215 |
4.619 ± 0.444 | 10.167 ± 0.491 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.669 ± 0.267 | 3.351 ± 0.259 |
5.718 ± 0.634 | 4.033 ± 0.34 |
6.02 ± 0.452 | 7.762 ± 0.505 |
7.062 ± 0.504 | 6.134 ± 0.178 |
1.155 ± 0.181 | 2.991 ± 0.288 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
