
Candidatus Scalindua rubra
Taxonomy: cellular organisms; Bacteria; PVC group; Planctomycetes; Candidatus Brocadiae; Candidatus Brocadiales; Candidatus Brocadiaceae; Candidatus Scalindua
Average proteome isoelectric point is 7.02
Get precalculated fractions of proteins
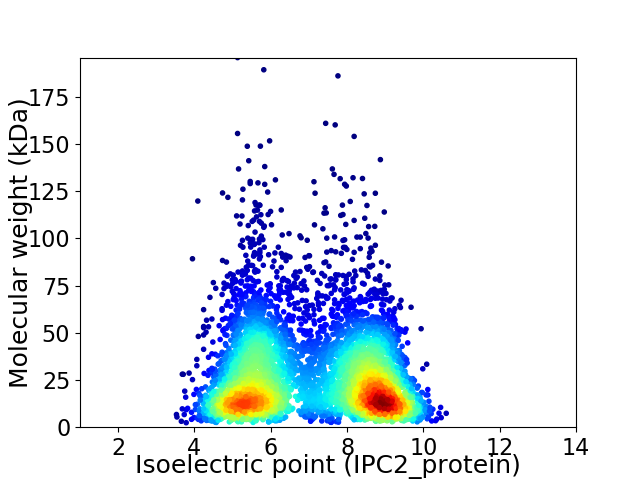
Virtual 2D-PAGE plot for 5207 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1E3X300|A0A1E3X300_9BACT Uncharacterized protein (Fragment) OS=Candidatus Scalindua rubra OX=1872076 GN=SCARUB_04923 PE=4 SV=1
MM1 pKa = 7.52NNNNLKK7 pKa = 10.34SCLILSIGVLLISTCEE23 pKa = 3.98EE24 pKa = 3.71PTEE27 pKa = 4.38PDD29 pKa = 3.18TTPPEE34 pKa = 4.17VTILDD39 pKa = 3.87PAEE42 pKa = 4.25GEE44 pKa = 4.33EE45 pKa = 4.04ISDD48 pKa = 3.78VFTVKK53 pKa = 9.48IDD55 pKa = 3.46ATDD58 pKa = 3.63DD59 pKa = 3.67NEE61 pKa = 4.06IEE63 pKa = 4.43KK64 pKa = 10.77VVLYY68 pKa = 10.37IDD70 pKa = 4.67FVPLDD75 pKa = 3.64SSLSEE80 pKa = 3.63KK81 pKa = 10.57DD82 pKa = 2.92IYY84 pKa = 11.24SFGWDD89 pKa = 3.31TDD91 pKa = 3.15IGDD94 pKa = 3.76NGEE97 pKa = 4.19YY98 pKa = 10.13TLLAQAFDD106 pKa = 3.45KK107 pKa = 11.26SGNKK111 pKa = 9.68SSSNPVNVTVINHH124 pKa = 7.15RR125 pKa = 11.84ILKK128 pKa = 9.13FVNTTWDD135 pKa = 4.35DD136 pKa = 3.05IDD138 pKa = 4.49FEE140 pKa = 4.51IAGVSDD146 pKa = 3.73VLLSLDD152 pKa = 4.02SIEE155 pKa = 5.21VEE157 pKa = 3.82IPKK160 pKa = 10.8NNGITNFYY168 pKa = 9.24GYY170 pKa = 10.18SWTGCGISIAWDD182 pKa = 3.3FDD184 pKa = 3.81IEE186 pKa = 4.85VGDD189 pKa = 3.99EE190 pKa = 3.88DD191 pKa = 5.36LRR193 pKa = 11.84WTVWVNPGAFYY204 pKa = 11.19LFLSNQSSVDD214 pKa = 3.1INYY217 pKa = 9.54VVVNKK222 pKa = 10.5DD223 pKa = 3.34LTDD226 pKa = 4.02LEE228 pKa = 4.94SWCYY232 pKa = 9.86WDD234 pKa = 4.12VPNDD238 pKa = 3.94GLVHH242 pKa = 6.59KK243 pKa = 9.9MGYY246 pKa = 8.86YY247 pKa = 9.29WADD250 pKa = 2.92
MM1 pKa = 7.52NNNNLKK7 pKa = 10.34SCLILSIGVLLISTCEE23 pKa = 3.98EE24 pKa = 3.71PTEE27 pKa = 4.38PDD29 pKa = 3.18TTPPEE34 pKa = 4.17VTILDD39 pKa = 3.87PAEE42 pKa = 4.25GEE44 pKa = 4.33EE45 pKa = 4.04ISDD48 pKa = 3.78VFTVKK53 pKa = 9.48IDD55 pKa = 3.46ATDD58 pKa = 3.63DD59 pKa = 3.67NEE61 pKa = 4.06IEE63 pKa = 4.43KK64 pKa = 10.77VVLYY68 pKa = 10.37IDD70 pKa = 4.67FVPLDD75 pKa = 3.64SSLSEE80 pKa = 3.63KK81 pKa = 10.57DD82 pKa = 2.92IYY84 pKa = 11.24SFGWDD89 pKa = 3.31TDD91 pKa = 3.15IGDD94 pKa = 3.76NGEE97 pKa = 4.19YY98 pKa = 10.13TLLAQAFDD106 pKa = 3.45KK107 pKa = 11.26SGNKK111 pKa = 9.68SSSNPVNVTVINHH124 pKa = 7.15RR125 pKa = 11.84ILKK128 pKa = 9.13FVNTTWDD135 pKa = 4.35DD136 pKa = 3.05IDD138 pKa = 4.49FEE140 pKa = 4.51IAGVSDD146 pKa = 3.73VLLSLDD152 pKa = 4.02SIEE155 pKa = 5.21VEE157 pKa = 3.82IPKK160 pKa = 10.8NNGITNFYY168 pKa = 9.24GYY170 pKa = 10.18SWTGCGISIAWDD182 pKa = 3.3FDD184 pKa = 3.81IEE186 pKa = 4.85VGDD189 pKa = 3.99EE190 pKa = 3.88DD191 pKa = 5.36LRR193 pKa = 11.84WTVWVNPGAFYY204 pKa = 11.19LFLSNQSSVDD214 pKa = 3.1INYY217 pKa = 9.54VVVNKK222 pKa = 10.5DD223 pKa = 3.34LTDD226 pKa = 4.02LEE228 pKa = 4.94SWCYY232 pKa = 9.86WDD234 pKa = 4.12VPNDD238 pKa = 3.94GLVHH242 pKa = 6.59KK243 pKa = 9.9MGYY246 pKa = 8.86YY247 pKa = 9.29WADD250 pKa = 2.92
Molecular weight: 28.06 kDa
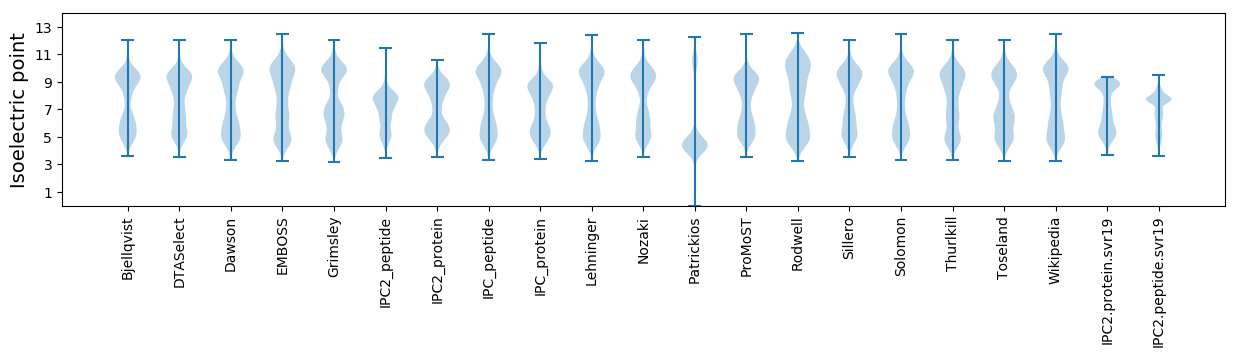
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1E3XE36|A0A1E3XE36_9BACT Methionyl-tRNA formyltransferase OS=Candidatus Scalindua rubra OX=1872076 GN=SCARUB_00954 PE=4 SV=1
MM1 pKa = 7.61KK2 pKa = 10.19IIAQRR7 pKa = 11.84SRR9 pKa = 11.84NQNSFNVKK17 pKa = 7.25TQSSKK22 pKa = 10.71EE23 pKa = 3.5IRR25 pKa = 11.84TGVNVRR31 pKa = 11.84ITIFAYY37 pKa = 7.29QTPVCVRR44 pKa = 11.84LRR46 pKa = 11.84TGRR49 pKa = 11.84HH50 pKa = 4.73GGEE53 pKa = 3.98NLKK56 pKa = 10.61VPLQFAIVVRR66 pKa = 11.84LEE68 pKa = 4.1GVNLFYY74 pKa = 11.08DD75 pKa = 3.82LGEE78 pKa = 4.12IKK80 pKa = 10.84GDD82 pKa = 3.32GRR84 pKa = 11.84SANGG88 pKa = 3.16
MM1 pKa = 7.61KK2 pKa = 10.19IIAQRR7 pKa = 11.84SRR9 pKa = 11.84NQNSFNVKK17 pKa = 7.25TQSSKK22 pKa = 10.71EE23 pKa = 3.5IRR25 pKa = 11.84TGVNVRR31 pKa = 11.84ITIFAYY37 pKa = 7.29QTPVCVRR44 pKa = 11.84LRR46 pKa = 11.84TGRR49 pKa = 11.84HH50 pKa = 4.73GGEE53 pKa = 3.98NLKK56 pKa = 10.61VPLQFAIVVRR66 pKa = 11.84LEE68 pKa = 4.1GVNLFYY74 pKa = 11.08DD75 pKa = 3.82LGEE78 pKa = 4.12IKK80 pKa = 10.84GDD82 pKa = 3.32GRR84 pKa = 11.84SANGG88 pKa = 3.16
Molecular weight: 9.83 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1364589 |
21 |
1692 |
262.1 |
29.77 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.767 ± 0.032 | 1.274 ± 0.015 |
5.498 ± 0.025 | 7.301 ± 0.037 |
4.588 ± 0.027 | 6.341 ± 0.033 |
1.96 ± 0.014 | 8.739 ± 0.035 |
8.461 ± 0.04 | 9.334 ± 0.033 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.317 ± 0.016 | 4.897 ± 0.026 |
3.582 ± 0.021 | 2.809 ± 0.018 |
4.893 ± 0.025 | 6.206 ± 0.027 |
5.019 ± 0.025 | 6.277 ± 0.027 |
1.033 ± 0.012 | 3.705 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
