
Human papillomavirus 122
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Betapapillomavirus; Betapapillomavirus 2
Average proteome isoelectric point is 6.01
Get precalculated fractions of proteins
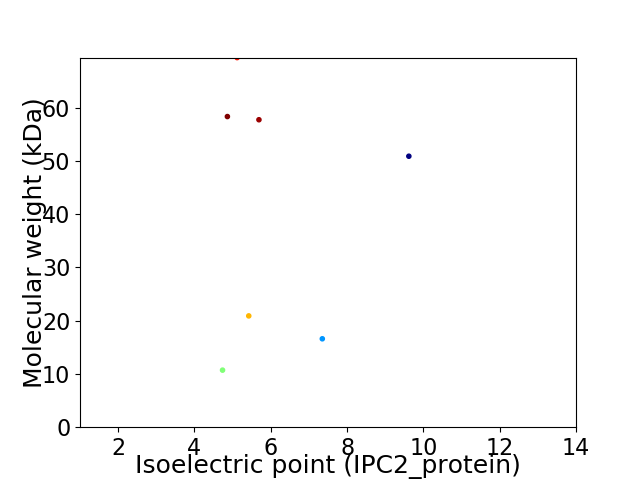
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
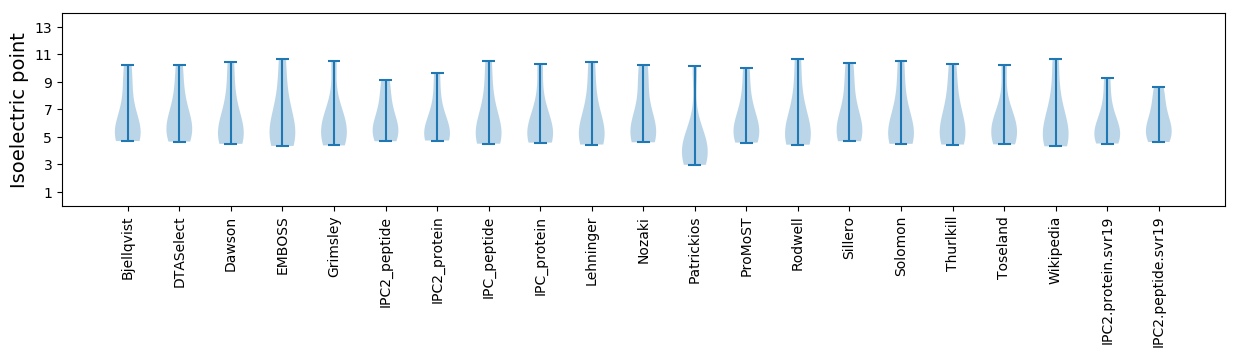
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D7P182|D7P182_9PAPI Replication protein E1 OS=Human papillomavirus 122 OX=765054 GN=E1 PE=3 SV=1
MM1 pKa = 7.6IGKK4 pKa = 8.88QPTIPDD10 pKa = 3.46VVLEE14 pKa = 4.15LQEE17 pKa = 4.85LVQPTDD23 pKa = 3.0LHH25 pKa = 7.68CYY27 pKa = 10.12EE28 pKa = 5.0EE29 pKa = 4.54LTEE32 pKa = 4.52EE33 pKa = 4.17PAEE36 pKa = 4.13EE37 pKa = 4.26EE38 pKa = 4.29QCLTPYY44 pKa = 10.38KK45 pKa = 10.38IIVACNCGTRR55 pKa = 11.84LRR57 pKa = 11.84LYY59 pKa = 10.95VFATDD64 pKa = 3.85LGIRR68 pKa = 11.84AQQEE72 pKa = 4.1LLLGEE77 pKa = 4.32VQLVCPEE84 pKa = 3.9CRR86 pKa = 11.84EE87 pKa = 3.77KK88 pKa = 10.94LRR90 pKa = 11.84RR91 pKa = 11.84HH92 pKa = 5.46EE93 pKa = 4.04
MM1 pKa = 7.6IGKK4 pKa = 8.88QPTIPDD10 pKa = 3.46VVLEE14 pKa = 4.15LQEE17 pKa = 4.85LVQPTDD23 pKa = 3.0LHH25 pKa = 7.68CYY27 pKa = 10.12EE28 pKa = 5.0EE29 pKa = 4.54LTEE32 pKa = 4.52EE33 pKa = 4.17PAEE36 pKa = 4.13EE37 pKa = 4.26EE38 pKa = 4.29QCLTPYY44 pKa = 10.38KK45 pKa = 10.38IIVACNCGTRR55 pKa = 11.84LRR57 pKa = 11.84LYY59 pKa = 10.95VFATDD64 pKa = 3.85LGIRR68 pKa = 11.84AQQEE72 pKa = 4.1LLLGEE77 pKa = 4.32VQLVCPEE84 pKa = 3.9CRR86 pKa = 11.84EE87 pKa = 3.77KK88 pKa = 10.94LRR90 pKa = 11.84RR91 pKa = 11.84HH92 pKa = 5.46EE93 pKa = 4.04
Molecular weight: 10.71 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D7P184|D7P184_9PAPI Uncharacterized protein E4 OS=Human papillomavirus 122 OX=765054 GN=E4 PE=4 SV=1
MM1 pKa = 7.66EE2 pKa = 5.01SLSEE6 pKa = 4.14RR7 pKa = 11.84FNALQEE13 pKa = 4.29TLMDD17 pKa = 4.67LYY19 pKa = 11.24EE20 pKa = 4.58SGRR23 pKa = 11.84EE24 pKa = 3.93DD25 pKa = 3.48LQSQIDD31 pKa = 3.25HH32 pKa = 6.24WQTLRR37 pKa = 11.84QEE39 pKa = 4.07QVLLYY44 pKa = 8.8FARR47 pKa = 11.84KK48 pKa = 8.52HH49 pKa = 4.52GVMRR53 pKa = 11.84LGYY56 pKa = 10.22QPVPPLATSEE66 pKa = 4.58SKK68 pKa = 11.11AKK70 pKa = 10.42DD71 pKa = 3.66AIGMVLLLQSLQKK84 pKa = 10.42SSYY87 pKa = 9.35GQEE90 pKa = 3.58PWTLVQTSLEE100 pKa = 4.48TVRR103 pKa = 11.84SPPANCFKK111 pKa = 10.8KK112 pKa = 10.31GPQNIEE118 pKa = 3.67VMFDD122 pKa = 3.5GDD124 pKa = 3.96PEE126 pKa = 5.54NIMSYY131 pKa = 7.43TVWTYY136 pKa = 10.87IYY138 pKa = 10.27YY139 pKa = 8.22QTNNDD144 pKa = 2.67TWEE147 pKa = 4.16KK148 pKa = 11.19VEE150 pKa = 4.03GQVDD154 pKa = 3.85YY155 pKa = 11.2FGAYY159 pKa = 9.53YY160 pKa = 10.68FEE162 pKa = 5.01GTLKK166 pKa = 9.74TYY168 pKa = 10.3YY169 pKa = 10.23INFEE173 pKa = 3.61TDD175 pKa = 2.51AARR178 pKa = 11.84FSKK181 pKa = 8.53TNKK184 pKa = 9.23WEE186 pKa = 3.72VHH188 pKa = 4.32VNKK191 pKa = 10.35NIVFAPVTSSSPPTGDD207 pKa = 3.01GGEE210 pKa = 4.16ASSNTVSRR218 pKa = 11.84SRR220 pKa = 11.84SHH222 pKa = 6.67SPQLSAPTLSSVQQRR237 pKa = 11.84PLSTTSRR244 pKa = 11.84RR245 pKa = 11.84YY246 pKa = 9.32RR247 pKa = 11.84RR248 pKa = 11.84KK249 pKa = 9.88ASSPTSTRR257 pKa = 11.84QKK259 pKa = 9.94RR260 pKa = 11.84QRR262 pKa = 11.84QKK264 pKa = 9.17ATTRR268 pKa = 11.84RR269 pKa = 11.84SRR271 pKa = 11.84SRR273 pKa = 11.84SRR275 pKa = 11.84GRR277 pKa = 11.84TATEE281 pKa = 3.51RR282 pKa = 11.84GGRR285 pKa = 11.84GGRR288 pKa = 11.84ASSADD293 pKa = 3.5SDD295 pKa = 4.01SSTGRR300 pKa = 11.84RR301 pKa = 11.84RR302 pKa = 11.84GRR304 pKa = 11.84GGGRR308 pKa = 11.84GPTTRR313 pKa = 11.84SQSRR317 pKa = 11.84SRR319 pKa = 11.84SRR321 pKa = 11.84SRR323 pKa = 11.84SRR325 pKa = 11.84SQARR329 pKa = 11.84GSSTRR334 pKa = 11.84GGIAPASVGASVRR347 pKa = 11.84SVGRR351 pKa = 11.84EE352 pKa = 3.3HH353 pKa = 7.29HH354 pKa = 6.46GRR356 pKa = 11.84LEE358 pKa = 3.98RR359 pKa = 11.84LLAEE363 pKa = 4.55AQDD366 pKa = 3.93PPIVILRR373 pKa = 11.84GEE375 pKa = 4.18ANVLKK380 pKa = 10.18CYY382 pKa = 10.28RR383 pKa = 11.84YY384 pKa = 9.27RR385 pKa = 11.84ARR387 pKa = 11.84KK388 pKa = 8.56RR389 pKa = 11.84HH390 pKa = 5.73LGLVKK395 pKa = 10.46YY396 pKa = 10.85YY397 pKa = 9.25STTWSWVGGDD407 pKa = 3.25CCDD410 pKa = 2.91RR411 pKa = 11.84VGRR414 pKa = 11.84SRR416 pKa = 11.84MLLAFDD422 pKa = 4.07TYY424 pKa = 10.68KK425 pKa = 10.55QRR427 pKa = 11.84EE428 pKa = 4.05QFITTMKK435 pKa = 10.51LPPNVDD441 pKa = 2.49WSFGHH446 pKa = 7.68LDD448 pKa = 3.65DD449 pKa = 5.58LL450 pKa = 5.21
MM1 pKa = 7.66EE2 pKa = 5.01SLSEE6 pKa = 4.14RR7 pKa = 11.84FNALQEE13 pKa = 4.29TLMDD17 pKa = 4.67LYY19 pKa = 11.24EE20 pKa = 4.58SGRR23 pKa = 11.84EE24 pKa = 3.93DD25 pKa = 3.48LQSQIDD31 pKa = 3.25HH32 pKa = 6.24WQTLRR37 pKa = 11.84QEE39 pKa = 4.07QVLLYY44 pKa = 8.8FARR47 pKa = 11.84KK48 pKa = 8.52HH49 pKa = 4.52GVMRR53 pKa = 11.84LGYY56 pKa = 10.22QPVPPLATSEE66 pKa = 4.58SKK68 pKa = 11.11AKK70 pKa = 10.42DD71 pKa = 3.66AIGMVLLLQSLQKK84 pKa = 10.42SSYY87 pKa = 9.35GQEE90 pKa = 3.58PWTLVQTSLEE100 pKa = 4.48TVRR103 pKa = 11.84SPPANCFKK111 pKa = 10.8KK112 pKa = 10.31GPQNIEE118 pKa = 3.67VMFDD122 pKa = 3.5GDD124 pKa = 3.96PEE126 pKa = 5.54NIMSYY131 pKa = 7.43TVWTYY136 pKa = 10.87IYY138 pKa = 10.27YY139 pKa = 8.22QTNNDD144 pKa = 2.67TWEE147 pKa = 4.16KK148 pKa = 11.19VEE150 pKa = 4.03GQVDD154 pKa = 3.85YY155 pKa = 11.2FGAYY159 pKa = 9.53YY160 pKa = 10.68FEE162 pKa = 5.01GTLKK166 pKa = 9.74TYY168 pKa = 10.3YY169 pKa = 10.23INFEE173 pKa = 3.61TDD175 pKa = 2.51AARR178 pKa = 11.84FSKK181 pKa = 8.53TNKK184 pKa = 9.23WEE186 pKa = 3.72VHH188 pKa = 4.32VNKK191 pKa = 10.35NIVFAPVTSSSPPTGDD207 pKa = 3.01GGEE210 pKa = 4.16ASSNTVSRR218 pKa = 11.84SRR220 pKa = 11.84SHH222 pKa = 6.67SPQLSAPTLSSVQQRR237 pKa = 11.84PLSTTSRR244 pKa = 11.84RR245 pKa = 11.84YY246 pKa = 9.32RR247 pKa = 11.84RR248 pKa = 11.84KK249 pKa = 9.88ASSPTSTRR257 pKa = 11.84QKK259 pKa = 9.94RR260 pKa = 11.84QRR262 pKa = 11.84QKK264 pKa = 9.17ATTRR268 pKa = 11.84RR269 pKa = 11.84SRR271 pKa = 11.84SRR273 pKa = 11.84SRR275 pKa = 11.84GRR277 pKa = 11.84TATEE281 pKa = 3.51RR282 pKa = 11.84GGRR285 pKa = 11.84GGRR288 pKa = 11.84ASSADD293 pKa = 3.5SDD295 pKa = 4.01SSTGRR300 pKa = 11.84RR301 pKa = 11.84RR302 pKa = 11.84GRR304 pKa = 11.84GGGRR308 pKa = 11.84GPTTRR313 pKa = 11.84SQSRR317 pKa = 11.84SRR319 pKa = 11.84SRR321 pKa = 11.84SRR323 pKa = 11.84SRR325 pKa = 11.84SQARR329 pKa = 11.84GSSTRR334 pKa = 11.84GGIAPASVGASVRR347 pKa = 11.84SVGRR351 pKa = 11.84EE352 pKa = 3.3HH353 pKa = 7.29HH354 pKa = 6.46GRR356 pKa = 11.84LEE358 pKa = 3.98RR359 pKa = 11.84LLAEE363 pKa = 4.55AQDD366 pKa = 3.93PPIVILRR373 pKa = 11.84GEE375 pKa = 4.18ANVLKK380 pKa = 10.18CYY382 pKa = 10.28RR383 pKa = 11.84YY384 pKa = 9.27RR385 pKa = 11.84ARR387 pKa = 11.84KK388 pKa = 8.56RR389 pKa = 11.84HH390 pKa = 5.73LGLVKK395 pKa = 10.46YY396 pKa = 10.85YY397 pKa = 9.25STTWSWVGGDD407 pKa = 3.25CCDD410 pKa = 2.91RR411 pKa = 11.84VGRR414 pKa = 11.84SRR416 pKa = 11.84MLLAFDD422 pKa = 4.07TYY424 pKa = 10.68KK425 pKa = 10.55QRR427 pKa = 11.84EE428 pKa = 4.05QFITTMKK435 pKa = 10.51LPPNVDD441 pKa = 2.49WSFGHH446 pKa = 7.68LDD448 pKa = 3.65DD449 pKa = 5.58LL450 pKa = 5.21
Molecular weight: 50.86 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2528 |
93 |
606 |
361.1 |
40.63 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.617 ± 0.311 | 2.057 ± 0.616 |
5.934 ± 0.435 | 6.646 ± 0.594 |
3.995 ± 0.592 | 6.843 ± 0.985 |
2.057 ± 0.167 | 4.786 ± 0.877 |
4.391 ± 0.634 | 8.703 ± 0.879 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.464 ± 0.282 | 3.837 ± 0.915 |
6.487 ± 1.546 | 5.498 ± 0.383 |
7.002 ± 1.048 | 7.595 ± 0.997 |
6.25 ± 0.673 | 6.29 ± 0.692 |
1.187 ± 0.304 | 3.362 ± 0.457 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
