
Tortoise microvirus 58
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 5.68
Get precalculated fractions of proteins
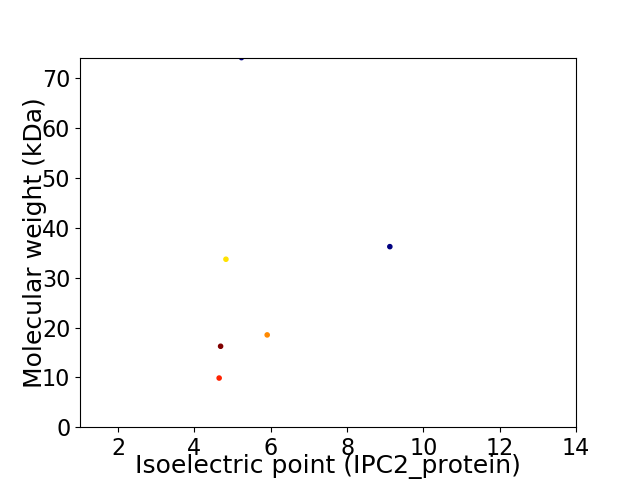
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
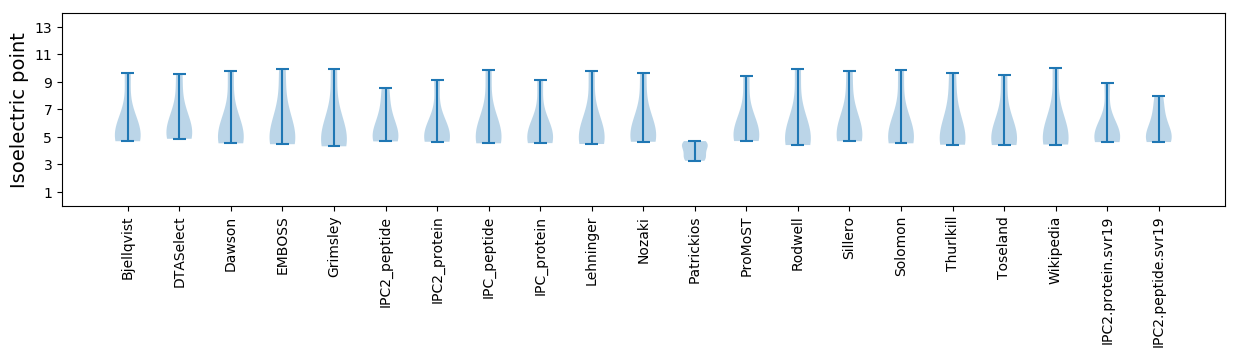
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W6S8|A0A4P8W6S8_9VIRU Major capsid protein OS=Tortoise microvirus 58 OX=2583163 PE=3 SV=1
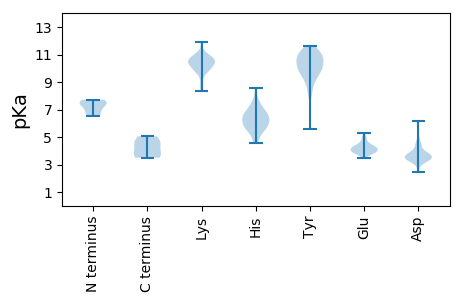
MM1 pKa = 7.72DD2 pKa = 3.39VAYY5 pKa = 10.15EE6 pKa = 4.4RR7 pKa = 11.84IHH9 pKa = 7.07GLTCAPNVALAIRR22 pKa = 11.84DD23 pKa = 3.41ISPTLKK29 pKa = 10.08RR30 pKa = 11.84VAPHH34 pKa = 6.51FLDD37 pKa = 4.85DD38 pKa = 3.66MEE40 pKa = 4.91LRR42 pKa = 11.84EE43 pKa = 4.24IGYY46 pKa = 9.96FDD48 pKa = 4.71SDD50 pKa = 4.7LEE52 pKa = 4.11FHH54 pKa = 6.66PTPAVVHH61 pKa = 6.09SWDD64 pKa = 3.66EE65 pKa = 4.23YY66 pKa = 11.26KK67 pKa = 11.07DD68 pKa = 3.51PEE70 pKa = 4.57QPVTPLSDD78 pKa = 3.42GQLKK82 pKa = 10.6DD83 pKa = 3.65FQRR86 pKa = 5.05
MM1 pKa = 7.72DD2 pKa = 3.39VAYY5 pKa = 10.15EE6 pKa = 4.4RR7 pKa = 11.84IHH9 pKa = 7.07GLTCAPNVALAIRR22 pKa = 11.84DD23 pKa = 3.41ISPTLKK29 pKa = 10.08RR30 pKa = 11.84VAPHH34 pKa = 6.51FLDD37 pKa = 4.85DD38 pKa = 3.66MEE40 pKa = 4.91LRR42 pKa = 11.84EE43 pKa = 4.24IGYY46 pKa = 9.96FDD48 pKa = 4.71SDD50 pKa = 4.7LEE52 pKa = 4.11FHH54 pKa = 6.66PTPAVVHH61 pKa = 6.09SWDD64 pKa = 3.66EE65 pKa = 4.23YY66 pKa = 11.26KK67 pKa = 11.07DD68 pKa = 3.51PEE70 pKa = 4.57QPVTPLSDD78 pKa = 3.42GQLKK82 pKa = 10.6DD83 pKa = 3.65FQRR86 pKa = 5.05
Molecular weight: 9.87 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W9U0|A0A4P8W9U0_9VIRU Uncharacterized protein OS=Tortoise microvirus 58 OX=2583163 PE=4 SV=1
MM1 pKa = 7.48ACSSPFFLEE10 pKa = 5.13RR11 pKa = 11.84ITDD14 pKa = 3.47YY15 pKa = 10.75RR16 pKa = 11.84GRR18 pKa = 11.84PVPLPCQKK26 pKa = 10.48CNCCRR31 pKa = 11.84ADD33 pKa = 3.12KK34 pKa = 8.35TTYY37 pKa = 5.55WTRR40 pKa = 11.84RR41 pKa = 11.84ANYY44 pKa = 7.99EE45 pKa = 3.66WTHH48 pKa = 5.57SVSSAFVTFTYY59 pKa = 10.51DD60 pKa = 3.98EE61 pKa = 4.06EE62 pKa = 4.28HH63 pKa = 6.63LPIGGSLEE71 pKa = 3.89PTLRR75 pKa = 11.84RR76 pKa = 11.84DD77 pKa = 3.48DD78 pKa = 3.47LHH80 pKa = 8.58RR81 pKa = 11.84YY82 pKa = 8.8IDD84 pKa = 3.93TVRR87 pKa = 11.84HH88 pKa = 5.62RR89 pKa = 11.84LKK91 pKa = 10.55KK92 pKa = 10.2DD93 pKa = 3.0GKK95 pKa = 9.74IPPRR99 pKa = 11.84CVPNFSYY106 pKa = 10.0MASGEE111 pKa = 4.22YY112 pKa = 10.24GDD114 pKa = 4.35SFGRR118 pKa = 11.84PHH120 pKa = 5.56YY121 pKa = 10.23HH122 pKa = 5.88VLFFGLDD129 pKa = 3.34FEE131 pKa = 4.83TCKK134 pKa = 10.69KK135 pKa = 9.93YY136 pKa = 10.81LKK138 pKa = 10.48KK139 pKa = 10.1SWPYY143 pKa = 11.12GSIAVNPVRR152 pKa = 11.84DD153 pKa = 3.36GAIRR157 pKa = 11.84YY158 pKa = 6.56VVSYY162 pKa = 10.45ISKK165 pKa = 9.33QLYY168 pKa = 9.89GEE170 pKa = 4.16EE171 pKa = 3.56RR172 pKa = 11.84DD173 pKa = 4.47ARR175 pKa = 11.84FFDD178 pKa = 4.3YY179 pKa = 10.55GVEE182 pKa = 4.1PPFLSVSRR190 pKa = 11.84GFGSGLFKK198 pKa = 10.98DD199 pKa = 3.44QAEE202 pKa = 4.51NIRR205 pKa = 11.84EE206 pKa = 3.92YY207 pKa = 11.27GALKK211 pKa = 10.35FGQKK215 pKa = 9.8FFSVPSYY222 pKa = 9.88WKK224 pKa = 10.44NKK226 pKa = 9.04YY227 pKa = 9.88IDD229 pKa = 3.65MDD231 pKa = 3.53IEE233 pKa = 4.34KK234 pKa = 10.46ADD236 pKa = 3.54EE237 pKa = 4.41RR238 pKa = 11.84YY239 pKa = 10.14EE240 pKa = 3.77RR241 pKa = 11.84AQRR244 pKa = 11.84LRR246 pKa = 11.84FEE248 pKa = 4.23RR249 pKa = 11.84WNLSGRR255 pKa = 11.84HH256 pKa = 4.65FQSPAGWTRR265 pKa = 11.84EE266 pKa = 3.82RR267 pKa = 11.84NRR269 pKa = 11.84VRR271 pKa = 11.84EE272 pKa = 3.85LVLYY276 pKa = 10.1QKK278 pKa = 10.85ARR280 pKa = 11.84NRR282 pKa = 11.84NSKK285 pKa = 9.87VGYY288 pKa = 8.26EE289 pKa = 4.05RR290 pKa = 11.84PIEE293 pKa = 3.94LRR295 pKa = 11.84PTGVGASIARR305 pKa = 11.84RR306 pKa = 11.84LRR308 pKa = 3.55
MM1 pKa = 7.48ACSSPFFLEE10 pKa = 5.13RR11 pKa = 11.84ITDD14 pKa = 3.47YY15 pKa = 10.75RR16 pKa = 11.84GRR18 pKa = 11.84PVPLPCQKK26 pKa = 10.48CNCCRR31 pKa = 11.84ADD33 pKa = 3.12KK34 pKa = 8.35TTYY37 pKa = 5.55WTRR40 pKa = 11.84RR41 pKa = 11.84ANYY44 pKa = 7.99EE45 pKa = 3.66WTHH48 pKa = 5.57SVSSAFVTFTYY59 pKa = 10.51DD60 pKa = 3.98EE61 pKa = 4.06EE62 pKa = 4.28HH63 pKa = 6.63LPIGGSLEE71 pKa = 3.89PTLRR75 pKa = 11.84RR76 pKa = 11.84DD77 pKa = 3.48DD78 pKa = 3.47LHH80 pKa = 8.58RR81 pKa = 11.84YY82 pKa = 8.8IDD84 pKa = 3.93TVRR87 pKa = 11.84HH88 pKa = 5.62RR89 pKa = 11.84LKK91 pKa = 10.55KK92 pKa = 10.2DD93 pKa = 3.0GKK95 pKa = 9.74IPPRR99 pKa = 11.84CVPNFSYY106 pKa = 10.0MASGEE111 pKa = 4.22YY112 pKa = 10.24GDD114 pKa = 4.35SFGRR118 pKa = 11.84PHH120 pKa = 5.56YY121 pKa = 10.23HH122 pKa = 5.88VLFFGLDD129 pKa = 3.34FEE131 pKa = 4.83TCKK134 pKa = 10.69KK135 pKa = 9.93YY136 pKa = 10.81LKK138 pKa = 10.48KK139 pKa = 10.1SWPYY143 pKa = 11.12GSIAVNPVRR152 pKa = 11.84DD153 pKa = 3.36GAIRR157 pKa = 11.84YY158 pKa = 6.56VVSYY162 pKa = 10.45ISKK165 pKa = 9.33QLYY168 pKa = 9.89GEE170 pKa = 4.16EE171 pKa = 3.56RR172 pKa = 11.84DD173 pKa = 4.47ARR175 pKa = 11.84FFDD178 pKa = 4.3YY179 pKa = 10.55GVEE182 pKa = 4.1PPFLSVSRR190 pKa = 11.84GFGSGLFKK198 pKa = 10.98DD199 pKa = 3.44QAEE202 pKa = 4.51NIRR205 pKa = 11.84EE206 pKa = 3.92YY207 pKa = 11.27GALKK211 pKa = 10.35FGQKK215 pKa = 9.8FFSVPSYY222 pKa = 9.88WKK224 pKa = 10.44NKK226 pKa = 9.04YY227 pKa = 9.88IDD229 pKa = 3.65MDD231 pKa = 3.53IEE233 pKa = 4.34KK234 pKa = 10.46ADD236 pKa = 3.54EE237 pKa = 4.41RR238 pKa = 11.84YY239 pKa = 10.14EE240 pKa = 3.77RR241 pKa = 11.84AQRR244 pKa = 11.84LRR246 pKa = 11.84FEE248 pKa = 4.23RR249 pKa = 11.84WNLSGRR255 pKa = 11.84HH256 pKa = 4.65FQSPAGWTRR265 pKa = 11.84EE266 pKa = 3.82RR267 pKa = 11.84NRR269 pKa = 11.84VRR271 pKa = 11.84EE272 pKa = 3.85LVLYY276 pKa = 10.1QKK278 pKa = 10.85ARR280 pKa = 11.84NRR282 pKa = 11.84NSKK285 pKa = 9.87VGYY288 pKa = 8.26EE289 pKa = 4.05RR290 pKa = 11.84PIEE293 pKa = 3.94LRR295 pKa = 11.84PTGVGASIARR305 pKa = 11.84RR306 pKa = 11.84LRR308 pKa = 3.55
Molecular weight: 36.24 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1677 |
86 |
668 |
279.5 |
31.46 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.977 ± 0.807 | 1.073 ± 0.55 |
7.036 ± 1.033 | 4.711 ± 0.835 |
4.949 ± 0.591 | 6.619 ± 0.619 |
1.67 ± 0.405 | 4.592 ± 0.457 |
4.055 ± 1.028 | 8.229 ± 0.423 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.087 ± 0.248 | 4.114 ± 0.588 |
4.949 ± 1.04 | 4.114 ± 0.703 |
6.559 ± 1.084 | 9.064 ± 0.749 |
6.082 ± 0.81 | 6.798 ± 0.586 |
1.491 ± 0.267 | 4.83 ± 0.915 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
