
Novosphingobium malaysiense
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Sphingomonadales; Sphingomonadaceae; Novosphingobium
Average proteome isoelectric point is 6.01
Get precalculated fractions of proteins
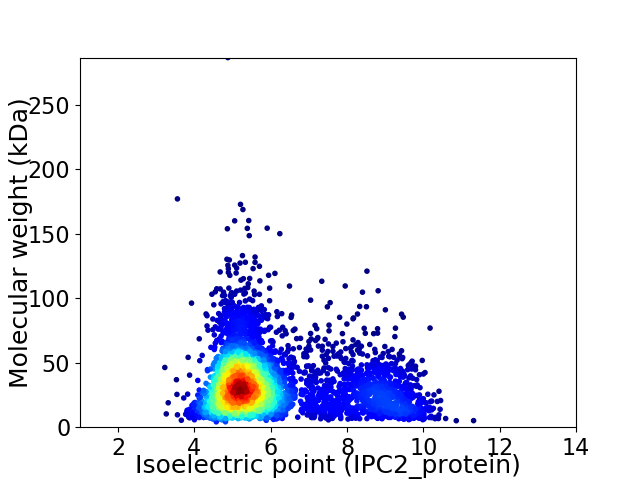
Virtual 2D-PAGE plot for 4270 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B1ZJH1|A0A0B1ZJH1_9SPHN Uncharacterized protein OS=Novosphingobium malaysiense OX=1348853 GN=LK12_10185 PE=4 SV=1
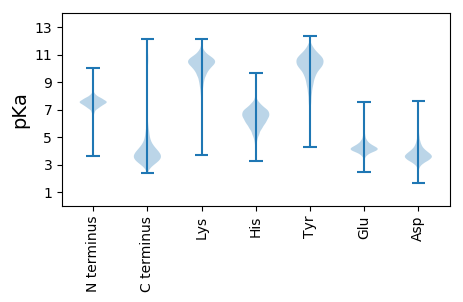
MM1 pKa = 7.4KK2 pKa = 10.17KK3 pKa = 10.08ISGLLLVGCSALALAGCGADD23 pKa = 5.0DD24 pKa = 4.29ISSPGAGSITINNPTPTPTPTPTPTSGVVTPAASCPSFNATGGLTDD70 pKa = 3.84SGTITGPEE78 pKa = 4.3GEE80 pKa = 4.38WRR82 pKa = 11.84VCTLPSLVDD91 pKa = 3.64VSSSLPKK98 pKa = 10.46VSGVLYY104 pKa = 10.17RR105 pKa = 11.84LNGRR109 pKa = 11.84VDD111 pKa = 3.49VGCDD115 pKa = 3.09GGFSAPTAAAPFATTTASCGGRR137 pKa = 11.84SLTADD142 pKa = 3.31TDD144 pKa = 3.71VTLTIDD150 pKa = 3.61PGVIVYY156 pKa = 9.83GEE158 pKa = 4.15NGAEE162 pKa = 4.11PAWLAVNRR170 pKa = 11.84GNKK173 pKa = 8.81INAVGTASQPIIFTSRR189 pKa = 11.84QNVVGTNDD197 pKa = 5.14DD198 pKa = 3.64SSQGQWGGVVLMGRR212 pKa = 11.84GIVTDD217 pKa = 3.95CAYY220 pKa = 10.72GSTAAGTCEE229 pKa = 3.94RR230 pKa = 11.84DD231 pKa = 3.55TEE233 pKa = 4.72GSANPAIFGGNDD245 pKa = 2.68NSYY248 pKa = 11.11NAGTMKK254 pKa = 10.33YY255 pKa = 9.56FQVRR259 pKa = 11.84YY260 pKa = 10.06SGFVLGANSEE270 pKa = 4.36LQSLTAEE277 pKa = 4.96GIGDD281 pKa = 4.12GTTLEE286 pKa = 4.48YY287 pKa = 10.22FQSVNSSDD295 pKa = 4.05DD296 pKa = 3.38GAEE299 pKa = 3.95FFGGKK304 pKa = 9.24VKK306 pKa = 10.57FKK308 pKa = 10.83HH309 pKa = 5.87YY310 pKa = 10.16IAVNADD316 pKa = 3.82DD317 pKa = 6.11DD318 pKa = 6.05SIDD321 pKa = 4.56LDD323 pKa = 4.04TGLEE327 pKa = 4.56GYY329 pKa = 9.16FQYY332 pKa = 11.62LLLLQRR338 pKa = 11.84DD339 pKa = 3.91GDD341 pKa = 3.95GDD343 pKa = 4.13ALMEE347 pKa = 4.69IDD349 pKa = 4.46SNGAEE354 pKa = 4.25SDD356 pKa = 3.6TPRR359 pKa = 11.84QKK361 pKa = 9.84TILANFTGVQPSDD374 pKa = 3.31NANSDD379 pKa = 3.56DD380 pKa = 3.8AAVLIRR386 pKa = 11.84GNSDD390 pKa = 2.9ITWANGIINVPNNEE404 pKa = 4.07CLRR407 pKa = 11.84LNGSGTVPATITADD421 pKa = 3.7SVVMTCNSAIYY432 pKa = 9.96LGTGTYY438 pKa = 8.26TAADD442 pKa = 3.7TEE444 pKa = 4.5AQFTAGTNNDD454 pKa = 3.49DD455 pKa = 4.09TFTSTLTSTFVNGSNEE471 pKa = 4.2SGVPAFDD478 pKa = 5.53ANTLDD483 pKa = 4.33SFFDD487 pKa = 3.4TTDD490 pKa = 3.33YY491 pKa = 11.11IGAVKK496 pKa = 10.35DD497 pKa = 5.16AADD500 pKa = 2.82TWYY503 pKa = 10.85RR504 pKa = 11.84GWTCDD509 pKa = 4.03NATADD514 pKa = 4.05FGSGSACTALPTTT527 pKa = 4.63
MM1 pKa = 7.4KK2 pKa = 10.17KK3 pKa = 10.08ISGLLLVGCSALALAGCGADD23 pKa = 5.0DD24 pKa = 4.29ISSPGAGSITINNPTPTPTPTPTPTSGVVTPAASCPSFNATGGLTDD70 pKa = 3.84SGTITGPEE78 pKa = 4.3GEE80 pKa = 4.38WRR82 pKa = 11.84VCTLPSLVDD91 pKa = 3.64VSSSLPKK98 pKa = 10.46VSGVLYY104 pKa = 10.17RR105 pKa = 11.84LNGRR109 pKa = 11.84VDD111 pKa = 3.49VGCDD115 pKa = 3.09GGFSAPTAAAPFATTTASCGGRR137 pKa = 11.84SLTADD142 pKa = 3.31TDD144 pKa = 3.71VTLTIDD150 pKa = 3.61PGVIVYY156 pKa = 9.83GEE158 pKa = 4.15NGAEE162 pKa = 4.11PAWLAVNRR170 pKa = 11.84GNKK173 pKa = 8.81INAVGTASQPIIFTSRR189 pKa = 11.84QNVVGTNDD197 pKa = 5.14DD198 pKa = 3.64SSQGQWGGVVLMGRR212 pKa = 11.84GIVTDD217 pKa = 3.95CAYY220 pKa = 10.72GSTAAGTCEE229 pKa = 3.94RR230 pKa = 11.84DD231 pKa = 3.55TEE233 pKa = 4.72GSANPAIFGGNDD245 pKa = 2.68NSYY248 pKa = 11.11NAGTMKK254 pKa = 10.33YY255 pKa = 9.56FQVRR259 pKa = 11.84YY260 pKa = 10.06SGFVLGANSEE270 pKa = 4.36LQSLTAEE277 pKa = 4.96GIGDD281 pKa = 4.12GTTLEE286 pKa = 4.48YY287 pKa = 10.22FQSVNSSDD295 pKa = 4.05DD296 pKa = 3.38GAEE299 pKa = 3.95FFGGKK304 pKa = 9.24VKK306 pKa = 10.57FKK308 pKa = 10.83HH309 pKa = 5.87YY310 pKa = 10.16IAVNADD316 pKa = 3.82DD317 pKa = 6.11DD318 pKa = 6.05SIDD321 pKa = 4.56LDD323 pKa = 4.04TGLEE327 pKa = 4.56GYY329 pKa = 9.16FQYY332 pKa = 11.62LLLLQRR338 pKa = 11.84DD339 pKa = 3.91GDD341 pKa = 3.95GDD343 pKa = 4.13ALMEE347 pKa = 4.69IDD349 pKa = 4.46SNGAEE354 pKa = 4.25SDD356 pKa = 3.6TPRR359 pKa = 11.84QKK361 pKa = 9.84TILANFTGVQPSDD374 pKa = 3.31NANSDD379 pKa = 3.56DD380 pKa = 3.8AAVLIRR386 pKa = 11.84GNSDD390 pKa = 2.9ITWANGIINVPNNEE404 pKa = 4.07CLRR407 pKa = 11.84LNGSGTVPATITADD421 pKa = 3.7SVVMTCNSAIYY432 pKa = 9.96LGTGTYY438 pKa = 8.26TAADD442 pKa = 3.7TEE444 pKa = 4.5AQFTAGTNNDD454 pKa = 3.49DD455 pKa = 4.09TFTSTLTSTFVNGSNEE471 pKa = 4.2SGVPAFDD478 pKa = 5.53ANTLDD483 pKa = 4.33SFFDD487 pKa = 3.4TTDD490 pKa = 3.33YY491 pKa = 11.11IGAVKK496 pKa = 10.35DD497 pKa = 5.16AADD500 pKa = 2.82TWYY503 pKa = 10.85RR504 pKa = 11.84GWTCDD509 pKa = 4.03NATADD514 pKa = 4.05FGSGSACTALPTTT527 pKa = 4.63
Molecular weight: 54.16 kDa
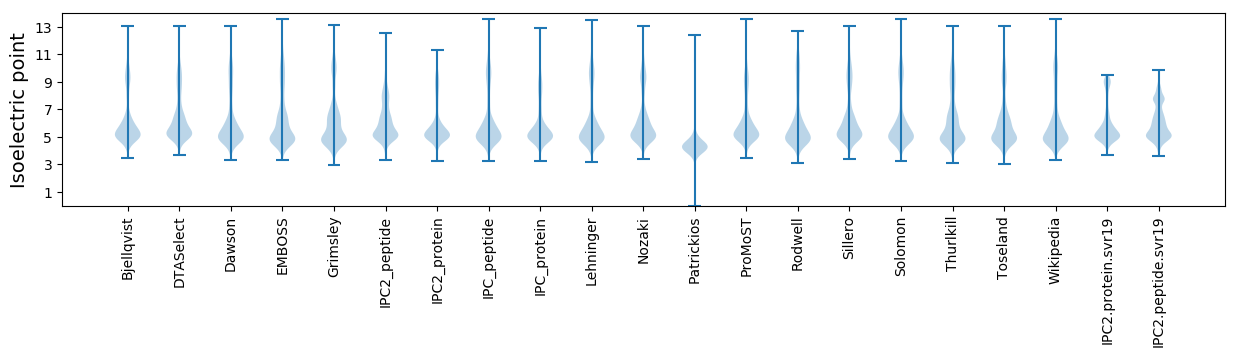
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B1ZVH2|A0A0B1ZVH2_9SPHN Uncharacterized protein OS=Novosphingobium malaysiense OX=1348853 GN=LK12_02185 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84LVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.54GFRR19 pKa = 11.84ARR21 pKa = 11.84TATAGGRR28 pKa = 11.84KK29 pKa = 8.05VLRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.02KK41 pKa = 10.61LSAA44 pKa = 4.03
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84LVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.54GFRR19 pKa = 11.84ARR21 pKa = 11.84TATAGGRR28 pKa = 11.84KK29 pKa = 8.05VLRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.02KK41 pKa = 10.61LSAA44 pKa = 4.03
Molecular weight: 5.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1374240 |
36 |
2676 |
321.8 |
34.94 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.327 ± 0.058 | 0.947 ± 0.012 |
6.067 ± 0.029 | 6.071 ± 0.031 |
3.654 ± 0.021 | 8.834 ± 0.032 |
2.103 ± 0.017 | 4.883 ± 0.028 |
3.291 ± 0.031 | 9.532 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.664 ± 0.02 | 2.662 ± 0.024 |
5.289 ± 0.027 | 3.101 ± 0.02 |
6.841 ± 0.036 | 5.429 ± 0.028 |
5.231 ± 0.029 | 7.219 ± 0.028 |
1.467 ± 0.014 | 2.386 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
