
Alstroemeria necrotic streak virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Ellioviricetes; Bunyavirales; Tospoviridae; Orthotospovirus; Alstroemeria necrotic streak orthotospovirus
Average proteome isoelectric point is 6.65
Get precalculated fractions of proteins
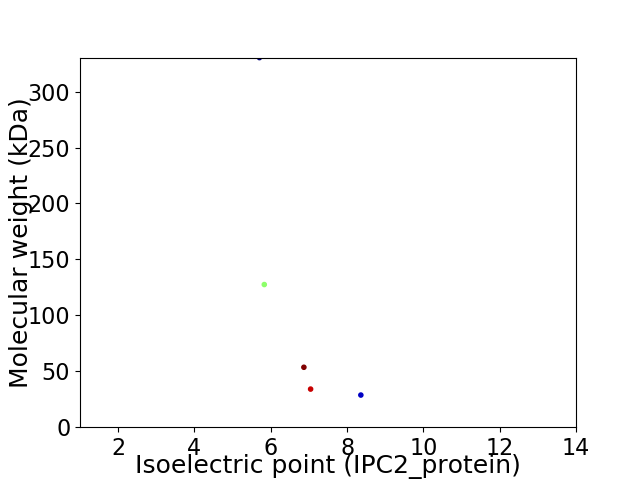
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
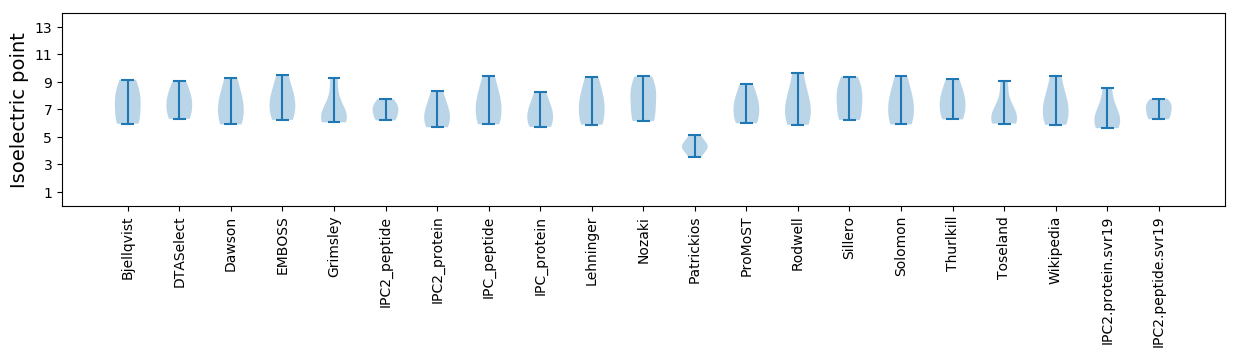
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3B8DZR5|A0A3B8DZR5_9VIRU Replicase OS=Alstroemeria necrotic streak virus OX=693450 PE=4 SV=1
MM1 pKa = 7.5KK2 pKa = 10.38LIKK5 pKa = 10.41LLEE8 pKa = 4.2LMVKK12 pKa = 10.55VSLFTIALSSILLSFLIFKK31 pKa = 9.23ATDD34 pKa = 3.11AKK36 pKa = 11.27VEE38 pKa = 4.54IIRR41 pKa = 11.84GDD43 pKa = 3.37HH44 pKa = 6.67PEE46 pKa = 4.52FYY48 pKa = 10.55DD49 pKa = 4.39DD50 pKa = 4.24SYY52 pKa = 12.03EE53 pKa = 4.18NEE55 pKa = 4.57LPVALPNQQKK65 pKa = 7.82TALEE69 pKa = 4.25TSTNLMLDD77 pKa = 3.37ASSIRR82 pKa = 11.84SRR84 pKa = 11.84QEE86 pKa = 3.24RR87 pKa = 11.84DD88 pKa = 3.06GKK90 pKa = 10.74LATASKK96 pKa = 10.5SDD98 pKa = 3.39SVTQKK103 pKa = 9.75TISVLDD109 pKa = 3.98LPNNCLNASLLKK121 pKa = 10.85CEE123 pKa = 4.29IKK125 pKa = 10.71GVSTYY130 pKa = 10.45NVYY133 pKa = 10.71YY134 pKa = 10.13QVEE137 pKa = 4.33NDD139 pKa = 3.66GVIYY143 pKa = 10.72SCVSDD148 pKa = 3.51TTEE151 pKa = 4.24SLGRR155 pKa = 11.84CDD157 pKa = 4.85NSQNLPKK164 pKa = 10.24KK165 pKa = 9.48FPEE168 pKa = 4.19VPVIPITRR176 pKa = 11.84LDD178 pKa = 3.75NKK180 pKa = 10.14RR181 pKa = 11.84HH182 pKa = 5.45FSVGTKK188 pKa = 9.6FFISEE193 pKa = 4.13SLAHH197 pKa = 6.34GNYY200 pKa = 9.39PITYY204 pKa = 9.75NSYY207 pKa = 8.05PTNGTVSLQTVKK219 pKa = 10.92LSGDD223 pKa = 3.73CKK225 pKa = 8.95ITKK228 pKa = 10.41SNFANPYY235 pKa = 7.71TVSITSPEE243 pKa = 4.14KK244 pKa = 10.56IMGYY248 pKa = 9.72LIKK251 pKa = 10.74KK252 pKa = 8.59PGEE255 pKa = 4.17DD256 pKa = 3.89AEE258 pKa = 4.6HH259 pKa = 6.44KK260 pKa = 10.38VIPFSGSTSITFSEE274 pKa = 4.16EE275 pKa = 3.91MLDD278 pKa = 3.68GDD280 pKa = 5.32HH281 pKa = 7.3NLLCGDD287 pKa = 4.55KK288 pKa = 10.39SAKK291 pKa = 9.51IPKK294 pKa = 7.78TNKK297 pKa = 9.47RR298 pKa = 11.84VRR300 pKa = 11.84DD301 pKa = 4.09CIIKK305 pKa = 9.92YY306 pKa = 10.28SKK308 pKa = 10.42SIYY311 pKa = 9.82KK312 pKa = 8.12QTACINFSWIRR323 pKa = 11.84LILIALLIYY332 pKa = 9.71FPIRR336 pKa = 11.84WLVNKK341 pKa = 6.89TTKK344 pKa = 10.24PLFLWYY350 pKa = 10.08DD351 pKa = 4.09LIGLITYY358 pKa = 7.77PILLILNYY366 pKa = 9.22LWKK369 pKa = 10.56YY370 pKa = 10.82FPFKK374 pKa = 10.67CSNCGNLCIVTHH386 pKa = 6.74EE387 pKa = 4.21CTKK390 pKa = 10.74RR391 pKa = 11.84CICNKK396 pKa = 9.61SKK398 pKa = 10.94ASRR401 pKa = 11.84EE402 pKa = 4.06HH403 pKa = 7.99SGEE406 pKa = 4.04CPILSKK412 pKa = 10.86EE413 pKa = 4.06ADD415 pKa = 3.41HH416 pKa = 7.0EE417 pKa = 4.6YY418 pKa = 10.82KK419 pKa = 9.81KK420 pKa = 10.43HH421 pKa = 5.83KK422 pKa = 6.45WTSMEE427 pKa = 3.86WFHH430 pKa = 7.65LIVNTKK436 pKa = 10.62LSLGLLKK443 pKa = 10.69FITEE447 pKa = 4.19LLIGLVILSQMPMSMAQTTQCLNGCFYY474 pKa = 11.52VPGCSYY480 pKa = 11.23LVTSKK485 pKa = 10.98FEE487 pKa = 3.99KK488 pKa = 10.67CPEE491 pKa = 4.05KK492 pKa = 10.71DD493 pKa = 2.88QCYY496 pKa = 10.9CNVQDD501 pKa = 4.94DD502 pKa = 4.99KK503 pKa = 11.2IIEE506 pKa = 4.67SIFGTNIVIEE516 pKa = 4.56GPNDD520 pKa = 3.95CIANQICSASSSIDD534 pKa = 3.41SLVKK538 pKa = 10.4CRR540 pKa = 11.84LGCEE544 pKa = 3.91YY545 pKa = 11.36LDD547 pKa = 4.03LFKK550 pKa = 11.16SKK552 pKa = 10.12PLYY555 pKa = 10.48KK556 pKa = 10.54GFPDD560 pKa = 3.47YY561 pKa = 10.88TGNSLGLTSVGLYY574 pKa = 5.74VTKK577 pKa = 10.65RR578 pKa = 11.84LRR580 pKa = 11.84NGIIDD585 pKa = 4.06SYY587 pKa = 11.43NGTDD591 pKa = 3.46RR592 pKa = 11.84ISGIVAGDD600 pKa = 3.71SLDD603 pKa = 4.57KK604 pKa = 11.24NDD606 pKa = 3.81TNIPEE611 pKa = 4.68NILPRR616 pKa = 11.84QSLIFDD622 pKa = 3.57STVDD626 pKa = 3.15GKK628 pKa = 10.53YY629 pKa = 10.28RR630 pKa = 11.84YY631 pKa = 9.28MIEE634 pKa = 3.85QALLGGGGTVFMLNDD649 pKa = 3.78KK650 pKa = 10.43TSGTAKK656 pKa = 10.58KK657 pKa = 8.37FTIYY661 pKa = 9.85IKK663 pKa = 10.66SVGVHH668 pKa = 5.02YY669 pKa = 10.32EE670 pKa = 3.8VAEE673 pKa = 4.53KK674 pKa = 9.97YY675 pKa = 5.9TTAPIQSTHH684 pKa = 5.09TDD686 pKa = 4.13FYY688 pKa = 10.5STCTGNCDD696 pKa = 3.29TCRR699 pKa = 11.84KK700 pKa = 8.9NQALTGFQDD709 pKa = 3.87FCITPTSYY717 pKa = 9.64WGCEE721 pKa = 3.99EE722 pKa = 4.13AWCFAINEE730 pKa = 4.33GATCGFCRR738 pKa = 11.84NVYY741 pKa = 11.08DD742 pKa = 3.24MDD744 pKa = 3.57KK745 pKa = 10.63SYY747 pKa = 11.12RR748 pKa = 11.84IYY750 pKa = 11.05SVLKK754 pKa = 7.88STIVADD760 pKa = 3.79VCVSGILGSHH770 pKa = 6.01CTRR773 pKa = 11.84ITEE776 pKa = 4.08EE777 pKa = 3.99VPYY780 pKa = 10.66EE781 pKa = 3.96NTIFQADD788 pKa = 3.53IQADD792 pKa = 3.72LHH794 pKa = 6.74NDD796 pKa = 4.41GITIGEE802 pKa = 4.79LIAHH806 pKa = 6.91GPDD809 pKa = 2.87SHH811 pKa = 8.27IYY813 pKa = 9.65SGNIANMNDD822 pKa = 3.43PVKK825 pKa = 10.48MFGHH829 pKa = 6.71PQLTHH834 pKa = 6.86DD835 pKa = 4.72GVPIFTKK842 pKa = 9.46KK843 pKa = 8.72TLEE846 pKa = 4.3GEE848 pKa = 4.15DD849 pKa = 4.8LSWDD853 pKa = 3.51CAAIGKK859 pKa = 10.08KK860 pKa = 10.03SVTIKK865 pKa = 9.08TCGYY869 pKa = 7.5DD870 pKa = 3.12TYY872 pKa = 10.89RR873 pKa = 11.84FKK875 pKa = 11.22SGLEE879 pKa = 3.96QISDD883 pKa = 3.4IPVIFKK889 pKa = 10.6DD890 pKa = 4.57FSSFFLEE897 pKa = 4.62KK898 pKa = 10.68SFNLGKK904 pKa = 10.72LKK906 pKa = 10.27MVVDD910 pKa = 5.14LPLDD914 pKa = 3.8LFKK917 pKa = 11.04VAPKK921 pKa = 10.12KK922 pKa = 10.48PSITSTRR929 pKa = 11.84LEE931 pKa = 3.94CSGCLLCGQGLSCNMEE947 pKa = 4.13FFSDD951 pKa = 3.92LTFSTAISIDD961 pKa = 3.6VCSLSTYY968 pKa = 10.46QLAIKK973 pKa = 10.13KK974 pKa = 10.17GSNKK978 pKa = 10.01YY979 pKa = 10.25NITMYY984 pKa = 10.77CSVNPDD990 pKa = 3.1KK991 pKa = 11.19KK992 pKa = 11.29KK993 pKa = 9.79MVLYY997 pKa = 10.32PEE999 pKa = 4.97GNPDD1003 pKa = 3.43LSVEE1007 pKa = 4.08VLINNVVVEE1016 pKa = 4.41EE1017 pKa = 4.55PEE1019 pKa = 4.55NIIDD1023 pKa = 4.1QNDD1026 pKa = 3.55EE1027 pKa = 4.09YY1028 pKa = 11.62AHH1030 pKa = 7.07EE1031 pKa = 4.17EE1032 pKa = 3.71QQYY1035 pKa = 11.29NSDD1038 pKa = 3.33SSAWSFWDD1046 pKa = 4.02YY1047 pKa = 11.1IKK1049 pKa = 11.3SPFNFIASYY1058 pKa = 10.19FGSFFDD1064 pKa = 4.2TIRR1067 pKa = 11.84VILLIAFIFLVVYY1080 pKa = 8.42FCSMLTSMCKK1090 pKa = 10.33GYY1092 pKa = 10.38IKK1094 pKa = 10.75NEE1096 pKa = 4.15SYY1098 pKa = 11.03KK1099 pKa = 10.98SRR1101 pKa = 11.84DD1102 pKa = 3.47KK1103 pKa = 11.84VEE1105 pKa = 5.44DD1106 pKa = 4.48DD1107 pKa = 5.11DD1108 pKa = 6.78DD1109 pKa = 6.19SEE1111 pKa = 4.65TTKK1114 pKa = 10.82PLLIKK1119 pKa = 10.22DD1120 pKa = 3.47TMTRR1124 pKa = 11.84RR1125 pKa = 11.84RR1126 pKa = 11.84PPMDD1130 pKa = 3.92FSHH1133 pKa = 6.98LVV1135 pKa = 3.07
MM1 pKa = 7.5KK2 pKa = 10.38LIKK5 pKa = 10.41LLEE8 pKa = 4.2LMVKK12 pKa = 10.55VSLFTIALSSILLSFLIFKK31 pKa = 9.23ATDD34 pKa = 3.11AKK36 pKa = 11.27VEE38 pKa = 4.54IIRR41 pKa = 11.84GDD43 pKa = 3.37HH44 pKa = 6.67PEE46 pKa = 4.52FYY48 pKa = 10.55DD49 pKa = 4.39DD50 pKa = 4.24SYY52 pKa = 12.03EE53 pKa = 4.18NEE55 pKa = 4.57LPVALPNQQKK65 pKa = 7.82TALEE69 pKa = 4.25TSTNLMLDD77 pKa = 3.37ASSIRR82 pKa = 11.84SRR84 pKa = 11.84QEE86 pKa = 3.24RR87 pKa = 11.84DD88 pKa = 3.06GKK90 pKa = 10.74LATASKK96 pKa = 10.5SDD98 pKa = 3.39SVTQKK103 pKa = 9.75TISVLDD109 pKa = 3.98LPNNCLNASLLKK121 pKa = 10.85CEE123 pKa = 4.29IKK125 pKa = 10.71GVSTYY130 pKa = 10.45NVYY133 pKa = 10.71YY134 pKa = 10.13QVEE137 pKa = 4.33NDD139 pKa = 3.66GVIYY143 pKa = 10.72SCVSDD148 pKa = 3.51TTEE151 pKa = 4.24SLGRR155 pKa = 11.84CDD157 pKa = 4.85NSQNLPKK164 pKa = 10.24KK165 pKa = 9.48FPEE168 pKa = 4.19VPVIPITRR176 pKa = 11.84LDD178 pKa = 3.75NKK180 pKa = 10.14RR181 pKa = 11.84HH182 pKa = 5.45FSVGTKK188 pKa = 9.6FFISEE193 pKa = 4.13SLAHH197 pKa = 6.34GNYY200 pKa = 9.39PITYY204 pKa = 9.75NSYY207 pKa = 8.05PTNGTVSLQTVKK219 pKa = 10.92LSGDD223 pKa = 3.73CKK225 pKa = 8.95ITKK228 pKa = 10.41SNFANPYY235 pKa = 7.71TVSITSPEE243 pKa = 4.14KK244 pKa = 10.56IMGYY248 pKa = 9.72LIKK251 pKa = 10.74KK252 pKa = 8.59PGEE255 pKa = 4.17DD256 pKa = 3.89AEE258 pKa = 4.6HH259 pKa = 6.44KK260 pKa = 10.38VIPFSGSTSITFSEE274 pKa = 4.16EE275 pKa = 3.91MLDD278 pKa = 3.68GDD280 pKa = 5.32HH281 pKa = 7.3NLLCGDD287 pKa = 4.55KK288 pKa = 10.39SAKK291 pKa = 9.51IPKK294 pKa = 7.78TNKK297 pKa = 9.47RR298 pKa = 11.84VRR300 pKa = 11.84DD301 pKa = 4.09CIIKK305 pKa = 9.92YY306 pKa = 10.28SKK308 pKa = 10.42SIYY311 pKa = 9.82KK312 pKa = 8.12QTACINFSWIRR323 pKa = 11.84LILIALLIYY332 pKa = 9.71FPIRR336 pKa = 11.84WLVNKK341 pKa = 6.89TTKK344 pKa = 10.24PLFLWYY350 pKa = 10.08DD351 pKa = 4.09LIGLITYY358 pKa = 7.77PILLILNYY366 pKa = 9.22LWKK369 pKa = 10.56YY370 pKa = 10.82FPFKK374 pKa = 10.67CSNCGNLCIVTHH386 pKa = 6.74EE387 pKa = 4.21CTKK390 pKa = 10.74RR391 pKa = 11.84CICNKK396 pKa = 9.61SKK398 pKa = 10.94ASRR401 pKa = 11.84EE402 pKa = 4.06HH403 pKa = 7.99SGEE406 pKa = 4.04CPILSKK412 pKa = 10.86EE413 pKa = 4.06ADD415 pKa = 3.41HH416 pKa = 7.0EE417 pKa = 4.6YY418 pKa = 10.82KK419 pKa = 9.81KK420 pKa = 10.43HH421 pKa = 5.83KK422 pKa = 6.45WTSMEE427 pKa = 3.86WFHH430 pKa = 7.65LIVNTKK436 pKa = 10.62LSLGLLKK443 pKa = 10.69FITEE447 pKa = 4.19LLIGLVILSQMPMSMAQTTQCLNGCFYY474 pKa = 11.52VPGCSYY480 pKa = 11.23LVTSKK485 pKa = 10.98FEE487 pKa = 3.99KK488 pKa = 10.67CPEE491 pKa = 4.05KK492 pKa = 10.71DD493 pKa = 2.88QCYY496 pKa = 10.9CNVQDD501 pKa = 4.94DD502 pKa = 4.99KK503 pKa = 11.2IIEE506 pKa = 4.67SIFGTNIVIEE516 pKa = 4.56GPNDD520 pKa = 3.95CIANQICSASSSIDD534 pKa = 3.41SLVKK538 pKa = 10.4CRR540 pKa = 11.84LGCEE544 pKa = 3.91YY545 pKa = 11.36LDD547 pKa = 4.03LFKK550 pKa = 11.16SKK552 pKa = 10.12PLYY555 pKa = 10.48KK556 pKa = 10.54GFPDD560 pKa = 3.47YY561 pKa = 10.88TGNSLGLTSVGLYY574 pKa = 5.74VTKK577 pKa = 10.65RR578 pKa = 11.84LRR580 pKa = 11.84NGIIDD585 pKa = 4.06SYY587 pKa = 11.43NGTDD591 pKa = 3.46RR592 pKa = 11.84ISGIVAGDD600 pKa = 3.71SLDD603 pKa = 4.57KK604 pKa = 11.24NDD606 pKa = 3.81TNIPEE611 pKa = 4.68NILPRR616 pKa = 11.84QSLIFDD622 pKa = 3.57STVDD626 pKa = 3.15GKK628 pKa = 10.53YY629 pKa = 10.28RR630 pKa = 11.84YY631 pKa = 9.28MIEE634 pKa = 3.85QALLGGGGTVFMLNDD649 pKa = 3.78KK650 pKa = 10.43TSGTAKK656 pKa = 10.58KK657 pKa = 8.37FTIYY661 pKa = 9.85IKK663 pKa = 10.66SVGVHH668 pKa = 5.02YY669 pKa = 10.32EE670 pKa = 3.8VAEE673 pKa = 4.53KK674 pKa = 9.97YY675 pKa = 5.9TTAPIQSTHH684 pKa = 5.09TDD686 pKa = 4.13FYY688 pKa = 10.5STCTGNCDD696 pKa = 3.29TCRR699 pKa = 11.84KK700 pKa = 8.9NQALTGFQDD709 pKa = 3.87FCITPTSYY717 pKa = 9.64WGCEE721 pKa = 3.99EE722 pKa = 4.13AWCFAINEE730 pKa = 4.33GATCGFCRR738 pKa = 11.84NVYY741 pKa = 11.08DD742 pKa = 3.24MDD744 pKa = 3.57KK745 pKa = 10.63SYY747 pKa = 11.12RR748 pKa = 11.84IYY750 pKa = 11.05SVLKK754 pKa = 7.88STIVADD760 pKa = 3.79VCVSGILGSHH770 pKa = 6.01CTRR773 pKa = 11.84ITEE776 pKa = 4.08EE777 pKa = 3.99VPYY780 pKa = 10.66EE781 pKa = 3.96NTIFQADD788 pKa = 3.53IQADD792 pKa = 3.72LHH794 pKa = 6.74NDD796 pKa = 4.41GITIGEE802 pKa = 4.79LIAHH806 pKa = 6.91GPDD809 pKa = 2.87SHH811 pKa = 8.27IYY813 pKa = 9.65SGNIANMNDD822 pKa = 3.43PVKK825 pKa = 10.48MFGHH829 pKa = 6.71PQLTHH834 pKa = 6.86DD835 pKa = 4.72GVPIFTKK842 pKa = 9.46KK843 pKa = 8.72TLEE846 pKa = 4.3GEE848 pKa = 4.15DD849 pKa = 4.8LSWDD853 pKa = 3.51CAAIGKK859 pKa = 10.08KK860 pKa = 10.03SVTIKK865 pKa = 9.08TCGYY869 pKa = 7.5DD870 pKa = 3.12TYY872 pKa = 10.89RR873 pKa = 11.84FKK875 pKa = 11.22SGLEE879 pKa = 3.96QISDD883 pKa = 3.4IPVIFKK889 pKa = 10.6DD890 pKa = 4.57FSSFFLEE897 pKa = 4.62KK898 pKa = 10.68SFNLGKK904 pKa = 10.72LKK906 pKa = 10.27MVVDD910 pKa = 5.14LPLDD914 pKa = 3.8LFKK917 pKa = 11.04VAPKK921 pKa = 10.12KK922 pKa = 10.48PSITSTRR929 pKa = 11.84LEE931 pKa = 3.94CSGCLLCGQGLSCNMEE947 pKa = 4.13FFSDD951 pKa = 3.92LTFSTAISIDD961 pKa = 3.6VCSLSTYY968 pKa = 10.46QLAIKK973 pKa = 10.13KK974 pKa = 10.17GSNKK978 pKa = 10.01YY979 pKa = 10.25NITMYY984 pKa = 10.77CSVNPDD990 pKa = 3.1KK991 pKa = 11.19KK992 pKa = 11.29KK993 pKa = 9.79MVLYY997 pKa = 10.32PEE999 pKa = 4.97GNPDD1003 pKa = 3.43LSVEE1007 pKa = 4.08VLINNVVVEE1016 pKa = 4.41EE1017 pKa = 4.55PEE1019 pKa = 4.55NIIDD1023 pKa = 4.1QNDD1026 pKa = 3.55EE1027 pKa = 4.09YY1028 pKa = 11.62AHH1030 pKa = 7.07EE1031 pKa = 4.17EE1032 pKa = 3.71QQYY1035 pKa = 11.29NSDD1038 pKa = 3.33SSAWSFWDD1046 pKa = 4.02YY1047 pKa = 11.1IKK1049 pKa = 11.3SPFNFIASYY1058 pKa = 10.19FGSFFDD1064 pKa = 4.2TIRR1067 pKa = 11.84VILLIAFIFLVVYY1080 pKa = 8.42FCSMLTSMCKK1090 pKa = 10.33GYY1092 pKa = 10.38IKK1094 pKa = 10.75NEE1096 pKa = 4.15SYY1098 pKa = 11.03KK1099 pKa = 10.98SRR1101 pKa = 11.84DD1102 pKa = 3.47KK1103 pKa = 11.84VEE1105 pKa = 5.44DD1106 pKa = 4.48DD1107 pKa = 5.11DD1108 pKa = 6.78DD1109 pKa = 6.19SEE1111 pKa = 4.65TTKK1114 pKa = 10.82PLLIKK1119 pKa = 10.22DD1120 pKa = 3.47TMTRR1124 pKa = 11.84RR1125 pKa = 11.84RR1126 pKa = 11.84PPMDD1130 pKa = 3.92FSHH1133 pKa = 6.98LVV1135 pKa = 3.07
Molecular weight: 127.53 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3B8DZQ8|A0A3B8DZQ8_9VIRU Envelopment polyprotein OS=Alstroemeria necrotic streak virus OX=693450 PE=3 SV=1
MM1 pKa = 7.54SKK3 pKa = 11.03AKK5 pKa = 8.99LTRR8 pKa = 11.84EE9 pKa = 3.98NIINLLTQSAEE20 pKa = 4.05VEE22 pKa = 4.25FEE24 pKa = 4.08EE25 pKa = 4.92EE26 pKa = 3.79QNQAAFNFQNFCQDD40 pKa = 3.11NLDD43 pKa = 3.96LVKK46 pKa = 10.75KK47 pKa = 9.99MSITSCLTFLKK58 pKa = 10.42NRR60 pKa = 11.84QSIMKK65 pKa = 9.42VIKK68 pKa = 10.61QSDD71 pKa = 3.74FTFGRR76 pKa = 11.84ITIKK80 pKa = 9.67KK81 pKa = 7.02TSEE84 pKa = 4.1RR85 pKa = 11.84IGATDD90 pKa = 3.11MTFRR94 pKa = 11.84RR95 pKa = 11.84LDD97 pKa = 3.09SMIRR101 pKa = 11.84VKK103 pKa = 10.81LIQEE107 pKa = 3.89TANAVNLDD115 pKa = 3.94AIKK118 pKa = 9.93TKK120 pKa = 10.22IASHH124 pKa = 6.91PLVQAYY130 pKa = 8.31GLPLNDD136 pKa = 3.59AKK138 pKa = 10.48SVRR141 pKa = 11.84LAIMLGGSIPLIASVDD157 pKa = 3.85SFEE160 pKa = 4.77MISVVLAIYY169 pKa = 10.44QDD171 pKa = 4.14AKK173 pKa = 11.24CKK175 pKa = 10.5DD176 pKa = 3.35LGINPKK182 pKa = 10.55KK183 pKa = 10.72YY184 pKa = 8.92DD185 pKa = 3.46TKK187 pKa = 10.19EE188 pKa = 3.85ALGKK192 pKa = 10.3VCTVLKK198 pKa = 10.79SKK200 pKa = 10.33GFSMDD205 pKa = 3.23DD206 pKa = 3.39TQVTKK211 pKa = 10.6GRR213 pKa = 11.84EE214 pKa = 3.67YY215 pKa = 11.16AAILSSCNPNAKK227 pKa = 9.97GSVAMEE233 pKa = 4.51HH234 pKa = 6.33YY235 pKa = 10.41SEE237 pKa = 4.88HH238 pKa = 6.42LNKK241 pKa = 9.79FYY243 pKa = 11.24EE244 pKa = 4.11MFGVKK249 pKa = 9.69KK250 pKa = 9.53EE251 pKa = 4.2SKK253 pKa = 10.02ISGVAA258 pKa = 2.97
MM1 pKa = 7.54SKK3 pKa = 11.03AKK5 pKa = 8.99LTRR8 pKa = 11.84EE9 pKa = 3.98NIINLLTQSAEE20 pKa = 4.05VEE22 pKa = 4.25FEE24 pKa = 4.08EE25 pKa = 4.92EE26 pKa = 3.79QNQAAFNFQNFCQDD40 pKa = 3.11NLDD43 pKa = 3.96LVKK46 pKa = 10.75KK47 pKa = 9.99MSITSCLTFLKK58 pKa = 10.42NRR60 pKa = 11.84QSIMKK65 pKa = 9.42VIKK68 pKa = 10.61QSDD71 pKa = 3.74FTFGRR76 pKa = 11.84ITIKK80 pKa = 9.67KK81 pKa = 7.02TSEE84 pKa = 4.1RR85 pKa = 11.84IGATDD90 pKa = 3.11MTFRR94 pKa = 11.84RR95 pKa = 11.84LDD97 pKa = 3.09SMIRR101 pKa = 11.84VKK103 pKa = 10.81LIQEE107 pKa = 3.89TANAVNLDD115 pKa = 3.94AIKK118 pKa = 9.93TKK120 pKa = 10.22IASHH124 pKa = 6.91PLVQAYY130 pKa = 8.31GLPLNDD136 pKa = 3.59AKK138 pKa = 10.48SVRR141 pKa = 11.84LAIMLGGSIPLIASVDD157 pKa = 3.85SFEE160 pKa = 4.77MISVVLAIYY169 pKa = 10.44QDD171 pKa = 4.14AKK173 pKa = 11.24CKK175 pKa = 10.5DD176 pKa = 3.35LGINPKK182 pKa = 10.55KK183 pKa = 10.72YY184 pKa = 8.92DD185 pKa = 3.46TKK187 pKa = 10.19EE188 pKa = 3.85ALGKK192 pKa = 10.3VCTVLKK198 pKa = 10.79SKK200 pKa = 10.33GFSMDD205 pKa = 3.23DD206 pKa = 3.39TQVTKK211 pKa = 10.6GRR213 pKa = 11.84EE214 pKa = 3.67YY215 pKa = 11.16AAILSSCNPNAKK227 pKa = 9.97GSVAMEE233 pKa = 4.51HH234 pKa = 6.33YY235 pKa = 10.41SEE237 pKa = 4.88HH238 pKa = 6.42LNKK241 pKa = 9.79FYY243 pKa = 11.24EE244 pKa = 4.11MFGVKK249 pKa = 9.69KK250 pKa = 9.53EE251 pKa = 4.2SKK253 pKa = 10.02ISGVAA258 pKa = 2.97
Molecular weight: 28.74 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5046 |
258 |
2874 |
1009.2 |
114.86 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.162 ± 0.518 | 2.279 ± 0.615 |
5.906 ± 0.297 | 6.817 ± 0.865 |
4.38 ± 0.254 | 4.261 ± 0.501 |
1.744 ± 0.186 | 8.125 ± 0.206 |
8.66 ± 0.356 | 9.81 ± 0.495 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.874 ± 0.381 | 6.223 ± 0.372 |
3.032 ± 0.57 | 2.893 ± 0.48 |
3.428 ± 0.379 | 9.176 ± 0.762 |
6.044 ± 0.333 | 5.371 ± 0.42 |
0.832 ± 0.188 | 3.904 ± 0.389 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
