
Fibrella aestuarina BUZ 2
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Cytophagia; Cytophagales; Cytophagaceae; Fibrella; Fibrella aestuarina
Average proteome isoelectric point is 6.91
Get precalculated fractions of proteins
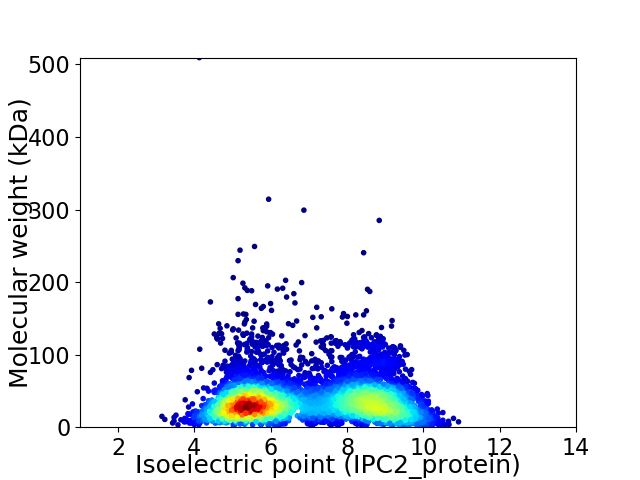
Virtual 2D-PAGE plot for 5627 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
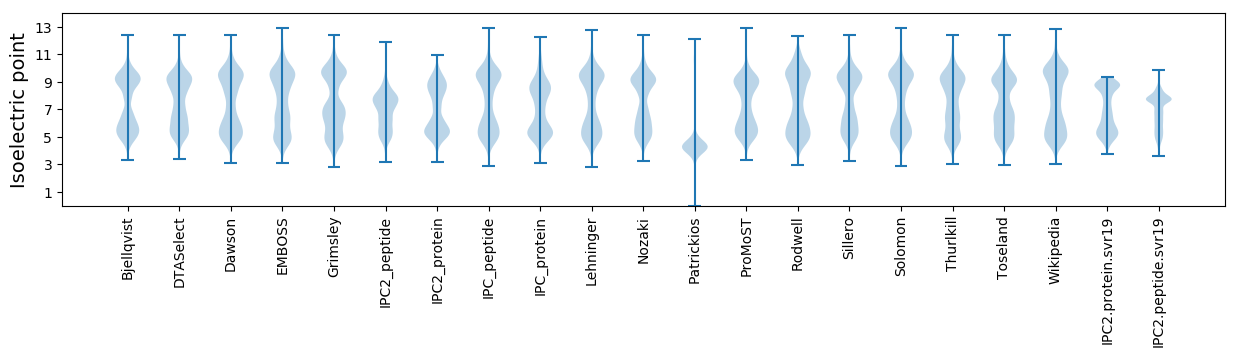
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|I0K1V9|I0K1V9_9BACT Transcriptional repressor CopY family OS=Fibrella aestuarina BUZ 2 OX=1166018 GN=FAES_0098 PE=3 SV=1
MM1 pKa = 7.49KK2 pKa = 10.3KK3 pKa = 10.6VILTGSALLAFVGAAWAQNSATLNQSGTYY32 pKa = 6.93QTAVQQQSGNNQVSTINQNQGAGVNTGNWAGTFQTGTVTNTATINQRR79 pKa = 11.84TGSQGNRR86 pKa = 11.84AAVGMVGGSGNVGTINQNGGALGLSGGATPPPGAGFDD123 pKa = 3.57QAVGDD128 pKa = 4.26GNFGGILQLGNTNTGVIINQNNLSQRR154 pKa = 11.84NTGEE158 pKa = 3.82IYY160 pKa = 10.81QNGDD164 pKa = 2.82NNANTTISQSNNSVGNQSMILQGFTSRR191 pKa = 11.84DD192 pKa = 3.29VASQVSSNNQATVEE206 pKa = 4.05QNGDD210 pKa = 3.05GTLAGSSSGNRR221 pKa = 11.84ADD223 pKa = 3.51VRR225 pKa = 11.84QLGEE229 pKa = 3.86QHH231 pKa = 7.05VSTTQQNGGSGGSSIGNTVSVDD253 pKa = 3.29QSGFGQTASVQQNGTANGSSTGNTATVVQSAFTNQATVEE292 pKa = 4.08QNGRR296 pKa = 11.84SEE298 pKa = 4.73GNTASVTQQGSTGGEE313 pKa = 4.04VPTLIGGGNTASVYY327 pKa = 10.5QNGGVAGLSAGNQATVSQDD346 pKa = 3.04GDD348 pKa = 3.43QHH350 pKa = 5.95TATVYY355 pKa = 10.66QGDD358 pKa = 4.11GTGGGTFSSEE368 pKa = 3.85ANTATVTQTGQTQTATIYY386 pKa = 8.43QQPGADD392 pKa = 3.67GSSALNTATITQSEE406 pKa = 4.81VEE408 pKa = 4.17NTATISQNNASVSNNATILQSGSASGVANNASIKK442 pKa = 10.8QNDD445 pKa = 3.67LSLGQTALVTQAGTNLFANVEE466 pKa = 4.33QNGITGGFTSSGNSATVTQTGDD488 pKa = 2.99FHH490 pKa = 7.0TVFAEE495 pKa = 4.07QNAGGNGGSINNQITITQVGVTGAHH520 pKa = 5.67NAEE523 pKa = 4.52VSQSDD528 pKa = 3.19ASGFNQITVDD538 pKa = 3.89QSGTSQQAFVNQSGVAFGIAAGSLGNQAVVTQSGTSNFADD578 pKa = 5.41LYY580 pKa = 9.59QTDD583 pKa = 3.71AAQDD587 pKa = 3.72GVITLTQSGNSNSAQLYY604 pKa = 8.19QVAGVTSNTATVEE617 pKa = 4.05QSGTTGIAAIQQIYY631 pKa = 10.86GSDD634 pKa = 3.34QANATILQSGTTNNAQIYY652 pKa = 8.78QYY654 pKa = 10.63QGTGNSVVIDD664 pKa = 3.64QLTTGTNNYY673 pKa = 10.19AEE675 pKa = 5.01IYY677 pKa = 10.39QGDD680 pKa = 4.22DD681 pKa = 2.73SFAPDD686 pKa = 3.45GSDD689 pKa = 3.58FNTTTVTQRR698 pKa = 11.84GTFNTVRR705 pKa = 11.84FDD707 pKa = 5.55LIGDD711 pKa = 3.81NNTATMSQTGRR722 pKa = 11.84GNLIRR727 pKa = 11.84GVAGTSPFSGYY738 pKa = 10.64AGQYY742 pKa = 10.68GNGNTLTITQVSDD755 pKa = 3.07AAGPGQTASVYY766 pKa = 9.96QAGNNNTTTITQTNNN781 pKa = 2.55
MM1 pKa = 7.49KK2 pKa = 10.3KK3 pKa = 10.6VILTGSALLAFVGAAWAQNSATLNQSGTYY32 pKa = 6.93QTAVQQQSGNNQVSTINQNQGAGVNTGNWAGTFQTGTVTNTATINQRR79 pKa = 11.84TGSQGNRR86 pKa = 11.84AAVGMVGGSGNVGTINQNGGALGLSGGATPPPGAGFDD123 pKa = 3.57QAVGDD128 pKa = 4.26GNFGGILQLGNTNTGVIINQNNLSQRR154 pKa = 11.84NTGEE158 pKa = 3.82IYY160 pKa = 10.81QNGDD164 pKa = 2.82NNANTTISQSNNSVGNQSMILQGFTSRR191 pKa = 11.84DD192 pKa = 3.29VASQVSSNNQATVEE206 pKa = 4.05QNGDD210 pKa = 3.05GTLAGSSSGNRR221 pKa = 11.84ADD223 pKa = 3.51VRR225 pKa = 11.84QLGEE229 pKa = 3.86QHH231 pKa = 7.05VSTTQQNGGSGGSSIGNTVSVDD253 pKa = 3.29QSGFGQTASVQQNGTANGSSTGNTATVVQSAFTNQATVEE292 pKa = 4.08QNGRR296 pKa = 11.84SEE298 pKa = 4.73GNTASVTQQGSTGGEE313 pKa = 4.04VPTLIGGGNTASVYY327 pKa = 10.5QNGGVAGLSAGNQATVSQDD346 pKa = 3.04GDD348 pKa = 3.43QHH350 pKa = 5.95TATVYY355 pKa = 10.66QGDD358 pKa = 4.11GTGGGTFSSEE368 pKa = 3.85ANTATVTQTGQTQTATIYY386 pKa = 8.43QQPGADD392 pKa = 3.67GSSALNTATITQSEE406 pKa = 4.81VEE408 pKa = 4.17NTATISQNNASVSNNATILQSGSASGVANNASIKK442 pKa = 10.8QNDD445 pKa = 3.67LSLGQTALVTQAGTNLFANVEE466 pKa = 4.33QNGITGGFTSSGNSATVTQTGDD488 pKa = 2.99FHH490 pKa = 7.0TVFAEE495 pKa = 4.07QNAGGNGGSINNQITITQVGVTGAHH520 pKa = 5.67NAEE523 pKa = 4.52VSQSDD528 pKa = 3.19ASGFNQITVDD538 pKa = 3.89QSGTSQQAFVNQSGVAFGIAAGSLGNQAVVTQSGTSNFADD578 pKa = 5.41LYY580 pKa = 9.59QTDD583 pKa = 3.71AAQDD587 pKa = 3.72GVITLTQSGNSNSAQLYY604 pKa = 8.19QVAGVTSNTATVEE617 pKa = 4.05QSGTTGIAAIQQIYY631 pKa = 10.86GSDD634 pKa = 3.34QANATILQSGTTNNAQIYY652 pKa = 8.78QYY654 pKa = 10.63QGTGNSVVIDD664 pKa = 3.64QLTTGTNNYY673 pKa = 10.19AEE675 pKa = 5.01IYY677 pKa = 10.39QGDD680 pKa = 4.22DD681 pKa = 2.73SFAPDD686 pKa = 3.45GSDD689 pKa = 3.58FNTTTVTQRR698 pKa = 11.84GTFNTVRR705 pKa = 11.84FDD707 pKa = 5.55LIGDD711 pKa = 3.81NNTATMSQTGRR722 pKa = 11.84GNLIRR727 pKa = 11.84GVAGTSPFSGYY738 pKa = 10.64AGQYY742 pKa = 10.68GNGNTLTITQVSDD755 pKa = 3.07AAGPGQTASVYY766 pKa = 9.96QAGNNNTTTITQTNNN781 pKa = 2.55
Molecular weight: 78.38 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|I0K8P0|I0K8P0_9BACT Hydrolase or acyltransferase (Alpha/beta hydrolase superfamily)-like protein OS=Fibrella aestuarina BUZ 2 OX=1166018 GN=FAES_2484 PE=4 SV=1
MM1 pKa = 7.32NVNTLTAPPRR11 pKa = 11.84SRR13 pKa = 11.84PAEE16 pKa = 3.84PVVTLHH22 pKa = 6.68RR23 pKa = 11.84PWLMPVLTGGLLLTSLLSVGVGAVAISPAEE53 pKa = 3.76VLAILGHH60 pKa = 5.4SLGLTGAVDD69 pKa = 3.38EE70 pKa = 4.72TKK72 pKa = 10.88AVILTGIRR80 pKa = 11.84LPRR83 pKa = 11.84VCLGLLIGAGLAVAGAAIQGLFRR106 pKa = 11.84NPLADD111 pKa = 4.41PGLIGISSGASLAAVGMIVLEE132 pKa = 4.03VTLFQKK138 pKa = 10.97LSGLLGFYY146 pKa = 10.63ALSLVAFGGACGTTLLVYY164 pKa = 10.55RR165 pKa = 11.84LARR168 pKa = 11.84VAGRR172 pKa = 11.84SVVTTMLLAGIAINALAGALTGLLTYY198 pKa = 10.68LATDD202 pKa = 3.53DD203 pKa = 3.71QLRR206 pKa = 11.84TITFWALGSLGGATWTTVLTILPFTIVVLLGLPRR240 pKa = 11.84LAKK243 pKa = 10.35SLNLLALGEE252 pKa = 4.37SQAAMLGVHH261 pKa = 5.24ITGLKK266 pKa = 9.71RR267 pKa = 11.84RR268 pKa = 11.84VIVLATLAVGTSVAVAGIIGFIGLVIPHH296 pKa = 7.81LIRR299 pKa = 11.84LVAGSDD305 pKa = 3.19HH306 pKa = 6.69RR307 pKa = 11.84RR308 pKa = 11.84VLTGSALGGAIVLTAADD325 pKa = 3.71ALARR329 pKa = 11.84TIVAPAEE336 pKa = 4.04LPIGILTALLGTPVFLWMLIKK357 pKa = 10.43EE358 pKa = 4.32RR359 pKa = 11.84STTT362 pKa = 3.4
MM1 pKa = 7.32NVNTLTAPPRR11 pKa = 11.84SRR13 pKa = 11.84PAEE16 pKa = 3.84PVVTLHH22 pKa = 6.68RR23 pKa = 11.84PWLMPVLTGGLLLTSLLSVGVGAVAISPAEE53 pKa = 3.76VLAILGHH60 pKa = 5.4SLGLTGAVDD69 pKa = 3.38EE70 pKa = 4.72TKK72 pKa = 10.88AVILTGIRR80 pKa = 11.84LPRR83 pKa = 11.84VCLGLLIGAGLAVAGAAIQGLFRR106 pKa = 11.84NPLADD111 pKa = 4.41PGLIGISSGASLAAVGMIVLEE132 pKa = 4.03VTLFQKK138 pKa = 10.97LSGLLGFYY146 pKa = 10.63ALSLVAFGGACGTTLLVYY164 pKa = 10.55RR165 pKa = 11.84LARR168 pKa = 11.84VAGRR172 pKa = 11.84SVVTTMLLAGIAINALAGALTGLLTYY198 pKa = 10.68LATDD202 pKa = 3.53DD203 pKa = 3.71QLRR206 pKa = 11.84TITFWALGSLGGATWTTVLTILPFTIVVLLGLPRR240 pKa = 11.84LAKK243 pKa = 10.35SLNLLALGEE252 pKa = 4.37SQAAMLGVHH261 pKa = 5.24ITGLKK266 pKa = 9.71RR267 pKa = 11.84RR268 pKa = 11.84VIVLATLAVGTSVAVAGIIGFIGLVIPHH296 pKa = 7.81LIRR299 pKa = 11.84LVAGSDD305 pKa = 3.19HH306 pKa = 6.69RR307 pKa = 11.84RR308 pKa = 11.84VLTGSALGGAIVLTAADD325 pKa = 3.71ALARR329 pKa = 11.84TIVAPAEE336 pKa = 4.04LPIGILTALLGTPVFLWMLIKK357 pKa = 10.43EE358 pKa = 4.32RR359 pKa = 11.84STTT362 pKa = 3.4
Molecular weight: 36.89 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2040039 |
29 |
4965 |
362.5 |
40.17 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.254 ± 0.037 | 0.747 ± 0.01 |
5.409 ± 0.026 | 4.748 ± 0.028 |
4.182 ± 0.023 | 7.436 ± 0.033 |
1.812 ± 0.018 | 5.096 ± 0.028 |
4.242 ± 0.031 | 10.357 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.054 ± 0.016 | 4.433 ± 0.029 |
4.959 ± 0.025 | 4.623 ± 0.025 |
5.851 ± 0.026 | 5.817 ± 0.032 |
6.859 ± 0.046 | 7.009 ± 0.024 |
1.362 ± 0.014 | 3.748 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
