
Devosia sp. Root436
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Devosiaceae; Devosia; unclassified Devosia
Average proteome isoelectric point is 6.22
Get precalculated fractions of proteins
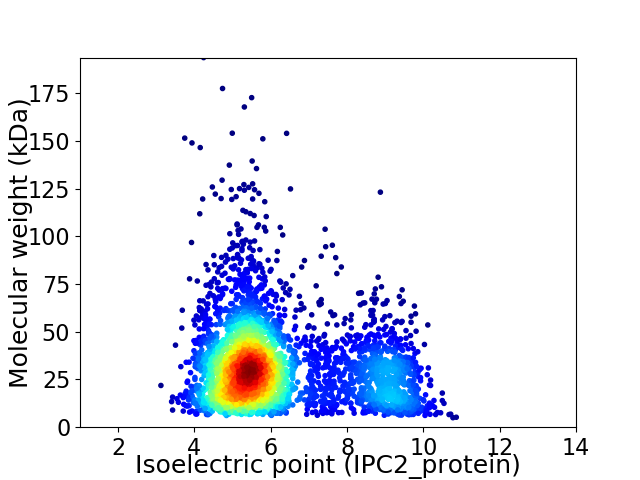
Virtual 2D-PAGE plot for 3690 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0Q7HW18|A0A0Q7HW18_9RHIZ GreA_GreB domain-containing protein OS=Devosia sp. Root436 OX=1736537 GN=ASD04_13455 PE=4 SV=1
MM1 pKa = 7.1THH3 pKa = 6.35FPCRR7 pKa = 11.84AAALAAGLMFSAGAMAQDD25 pKa = 3.63DD26 pKa = 4.56PGWHH30 pKa = 5.98GNAYY34 pKa = 10.65DD35 pKa = 3.91GMTTLFYY42 pKa = 10.77GVPQSDD48 pKa = 3.56HH49 pKa = 8.22VEE51 pKa = 3.9IALSCQAGSKK61 pKa = 9.66AANFVFAFAPIEE73 pKa = 4.19AVDD76 pKa = 4.18GVQVQVTLEE85 pKa = 4.2ADD87 pKa = 4.57DD88 pKa = 4.19ISLPIQTTGTLMQMDD103 pKa = 5.0DD104 pKa = 3.92LFLLEE109 pKa = 4.85GEE111 pKa = 4.63VAVDD115 pKa = 3.9ARR117 pKa = 11.84LIDD120 pKa = 4.13LLGSPGMLSVFVEE133 pKa = 4.9DD134 pKa = 5.06GAAEE138 pKa = 4.13YY139 pKa = 9.71PLEE142 pKa = 4.54GALDD146 pKa = 3.59AAAALIQACGQDD158 pKa = 3.61AEE160 pKa = 4.55TAAIEE165 pKa = 4.53SCDD168 pKa = 3.57LNAWIEE174 pKa = 4.26AEE176 pKa = 4.44GPAAQVIRR184 pKa = 11.84DD185 pKa = 4.05GPSANATAIADD196 pKa = 3.62MPQPYY201 pKa = 9.88EE202 pKa = 4.71GYY204 pKa = 10.77DD205 pKa = 3.24GMAYY209 pKa = 7.12PTASITGSRR218 pKa = 11.84DD219 pKa = 2.17GWFRR223 pKa = 11.84IKK225 pKa = 10.6EE226 pKa = 4.31VVTNLYY232 pKa = 10.95AEE234 pKa = 4.89ADD236 pKa = 3.78PVTVFSGEE244 pKa = 3.55GWVPGKK250 pKa = 10.59ALGLYY255 pKa = 9.67VEE257 pKa = 5.26SPALLDD263 pKa = 4.67RR264 pKa = 11.84PADD267 pKa = 4.0DD268 pKa = 4.13AAVAVDD274 pKa = 4.8FSDD277 pKa = 3.6GGEE280 pKa = 4.01NFLTVDD286 pKa = 3.67RR287 pKa = 11.84LYY289 pKa = 10.91DD290 pKa = 3.62CRR292 pKa = 11.84GDD294 pKa = 3.25WVEE297 pKa = 4.86IGGTYY302 pKa = 9.63AGKK305 pKa = 10.22RR306 pKa = 11.84VRR308 pKa = 11.84GWSSDD313 pKa = 2.82ICEE316 pKa = 4.32SQITTCSS323 pKa = 3.11
MM1 pKa = 7.1THH3 pKa = 6.35FPCRR7 pKa = 11.84AAALAAGLMFSAGAMAQDD25 pKa = 3.63DD26 pKa = 4.56PGWHH30 pKa = 5.98GNAYY34 pKa = 10.65DD35 pKa = 3.91GMTTLFYY42 pKa = 10.77GVPQSDD48 pKa = 3.56HH49 pKa = 8.22VEE51 pKa = 3.9IALSCQAGSKK61 pKa = 9.66AANFVFAFAPIEE73 pKa = 4.19AVDD76 pKa = 4.18GVQVQVTLEE85 pKa = 4.2ADD87 pKa = 4.57DD88 pKa = 4.19ISLPIQTTGTLMQMDD103 pKa = 5.0DD104 pKa = 3.92LFLLEE109 pKa = 4.85GEE111 pKa = 4.63VAVDD115 pKa = 3.9ARR117 pKa = 11.84LIDD120 pKa = 4.13LLGSPGMLSVFVEE133 pKa = 4.9DD134 pKa = 5.06GAAEE138 pKa = 4.13YY139 pKa = 9.71PLEE142 pKa = 4.54GALDD146 pKa = 3.59AAAALIQACGQDD158 pKa = 3.61AEE160 pKa = 4.55TAAIEE165 pKa = 4.53SCDD168 pKa = 3.57LNAWIEE174 pKa = 4.26AEE176 pKa = 4.44GPAAQVIRR184 pKa = 11.84DD185 pKa = 4.05GPSANATAIADD196 pKa = 3.62MPQPYY201 pKa = 9.88EE202 pKa = 4.71GYY204 pKa = 10.77DD205 pKa = 3.24GMAYY209 pKa = 7.12PTASITGSRR218 pKa = 11.84DD219 pKa = 2.17GWFRR223 pKa = 11.84IKK225 pKa = 10.6EE226 pKa = 4.31VVTNLYY232 pKa = 10.95AEE234 pKa = 4.89ADD236 pKa = 3.78PVTVFSGEE244 pKa = 3.55GWVPGKK250 pKa = 10.59ALGLYY255 pKa = 9.67VEE257 pKa = 5.26SPALLDD263 pKa = 4.67RR264 pKa = 11.84PADD267 pKa = 4.0DD268 pKa = 4.13AAVAVDD274 pKa = 4.8FSDD277 pKa = 3.6GGEE280 pKa = 4.01NFLTVDD286 pKa = 3.67RR287 pKa = 11.84LYY289 pKa = 10.91DD290 pKa = 3.62CRR292 pKa = 11.84GDD294 pKa = 3.25WVEE297 pKa = 4.86IGGTYY302 pKa = 9.63AGKK305 pKa = 10.22RR306 pKa = 11.84VRR308 pKa = 11.84GWSSDD313 pKa = 2.82ICEE316 pKa = 4.32SQITTCSS323 pKa = 3.11
Molecular weight: 34.02 kDa
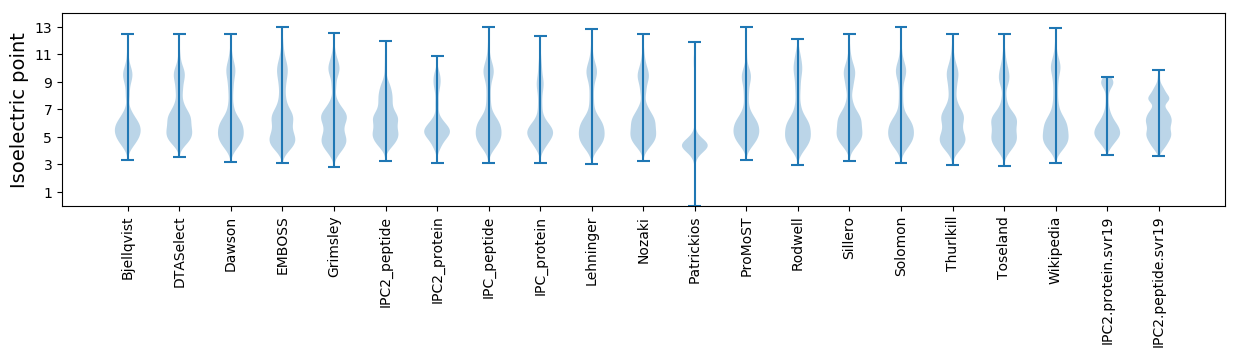
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0Q7I9G1|A0A0Q7I9G1_9RHIZ NepR domain-containing protein OS=Devosia sp. Root436 OX=1736537 GN=ASD04_06510 PE=4 SV=1
MM1 pKa = 6.92TADD4 pKa = 3.44VTRR7 pKa = 11.84LIAYY11 pKa = 7.31YY12 pKa = 10.01KK13 pKa = 10.52SPLGRR18 pKa = 11.84ISRR21 pKa = 11.84ALVRR25 pKa = 11.84EE26 pKa = 3.98QVLGLAGDD34 pKa = 3.83VAGKK38 pKa = 10.37RR39 pKa = 11.84ILGLGFATPYY49 pKa = 9.98MRR51 pKa = 11.84FTLEE55 pKa = 3.34RR56 pKa = 11.84AEE58 pKa = 4.01RR59 pKa = 11.84VLAFMPARR67 pKa = 11.84QGASAWPRR75 pKa = 11.84EE76 pKa = 4.64GPSHH80 pKa = 6.71TVLCDD85 pKa = 3.6PLEE88 pKa = 4.41MPLTDD93 pKa = 3.66AAIDD97 pKa = 3.65LTVAIHH103 pKa = 6.15ALEE106 pKa = 4.48HH107 pKa = 5.66VADD110 pKa = 4.16AEE112 pKa = 4.11EE113 pKa = 4.41LMRR116 pKa = 11.84EE117 pKa = 4.55LWRR120 pKa = 11.84ITAPNGHH127 pKa = 7.34LLLVVPRR134 pKa = 11.84RR135 pKa = 11.84RR136 pKa = 11.84GIWAQRR142 pKa = 11.84DD143 pKa = 3.6NTPFGSGNPYY153 pKa = 10.58SGGQLDD159 pKa = 3.95KK160 pKa = 11.17LLRR163 pKa = 11.84DD164 pKa = 3.48HH165 pKa = 7.11SFVPEE170 pKa = 3.97AWRR173 pKa = 11.84DD174 pKa = 3.74GLFLPPFQSSLVLKK188 pKa = 7.83STRR191 pKa = 11.84FFEE194 pKa = 4.35RR195 pKa = 11.84VGRR198 pKa = 11.84LFGPAMSGVICVRR211 pKa = 11.84ARR213 pKa = 11.84KK214 pKa = 8.57EE215 pKa = 3.76AFPAVPRR222 pKa = 11.84RR223 pKa = 11.84KK224 pKa = 9.36RR225 pKa = 11.84EE226 pKa = 3.69EE227 pKa = 3.76RR228 pKa = 11.84FVRR231 pKa = 11.84VPGLSTATARR241 pKa = 11.84SSWSKK246 pKa = 11.0GRR248 pKa = 11.84FPLSRR253 pKa = 11.84EE254 pKa = 3.88
MM1 pKa = 6.92TADD4 pKa = 3.44VTRR7 pKa = 11.84LIAYY11 pKa = 7.31YY12 pKa = 10.01KK13 pKa = 10.52SPLGRR18 pKa = 11.84ISRR21 pKa = 11.84ALVRR25 pKa = 11.84EE26 pKa = 3.98QVLGLAGDD34 pKa = 3.83VAGKK38 pKa = 10.37RR39 pKa = 11.84ILGLGFATPYY49 pKa = 9.98MRR51 pKa = 11.84FTLEE55 pKa = 3.34RR56 pKa = 11.84AEE58 pKa = 4.01RR59 pKa = 11.84VLAFMPARR67 pKa = 11.84QGASAWPRR75 pKa = 11.84EE76 pKa = 4.64GPSHH80 pKa = 6.71TVLCDD85 pKa = 3.6PLEE88 pKa = 4.41MPLTDD93 pKa = 3.66AAIDD97 pKa = 3.65LTVAIHH103 pKa = 6.15ALEE106 pKa = 4.48HH107 pKa = 5.66VADD110 pKa = 4.16AEE112 pKa = 4.11EE113 pKa = 4.41LMRR116 pKa = 11.84EE117 pKa = 4.55LWRR120 pKa = 11.84ITAPNGHH127 pKa = 7.34LLLVVPRR134 pKa = 11.84RR135 pKa = 11.84RR136 pKa = 11.84GIWAQRR142 pKa = 11.84DD143 pKa = 3.6NTPFGSGNPYY153 pKa = 10.58SGGQLDD159 pKa = 3.95KK160 pKa = 11.17LLRR163 pKa = 11.84DD164 pKa = 3.48HH165 pKa = 7.11SFVPEE170 pKa = 3.97AWRR173 pKa = 11.84DD174 pKa = 3.74GLFLPPFQSSLVLKK188 pKa = 7.83STRR191 pKa = 11.84FFEE194 pKa = 4.35RR195 pKa = 11.84VGRR198 pKa = 11.84LFGPAMSGVICVRR211 pKa = 11.84ARR213 pKa = 11.84KK214 pKa = 8.57EE215 pKa = 3.76AFPAVPRR222 pKa = 11.84RR223 pKa = 11.84KK224 pKa = 9.36RR225 pKa = 11.84EE226 pKa = 3.69EE227 pKa = 3.76RR228 pKa = 11.84FVRR231 pKa = 11.84VPGLSTATARR241 pKa = 11.84SSWSKK246 pKa = 11.0GRR248 pKa = 11.84FPLSRR253 pKa = 11.84EE254 pKa = 3.88
Molecular weight: 28.42 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1134231 |
41 |
1832 |
307.4 |
33.25 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.62 ± 0.058 | 0.669 ± 0.011 |
5.834 ± 0.03 | 5.511 ± 0.038 |
3.774 ± 0.029 | 8.672 ± 0.043 |
1.994 ± 0.019 | 5.537 ± 0.03 |
3.079 ± 0.034 | 10.286 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.595 ± 0.019 | 2.723 ± 0.024 |
5.023 ± 0.031 | 3.167 ± 0.021 |
6.569 ± 0.041 | 5.31 ± 0.026 |
5.45 ± 0.026 | 7.602 ± 0.032 |
1.313 ± 0.018 | 2.271 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
