
Candidatus Altiarchaeales archaeon WOR_SM1_SCG
Taxonomy: cellular organisms; Archaea; DPANN group; Candidatus Altiarchaeota; Candidatus Altiarchaeales; unclassified Candidatus Altiarchaeales
Average proteome isoelectric point is 6.73
Get precalculated fractions of proteins
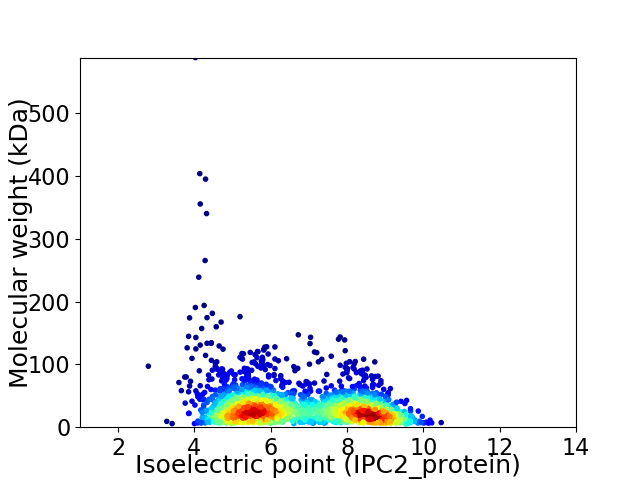
Virtual 2D-PAGE plot for 2164 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1D2RA85|A0A1D2RA85_9ARCH INTEIN_C_TER domain-containing protein (Fragment) OS=Candidatus Altiarchaeales archaeon WOR_SM1_SCG OX=1849261 GN=BEH94_04500 PE=3 SV=1
MM1 pKa = 6.85KK2 pKa = 9.63TSNIILLIFVFEE14 pKa = 4.97IILSNLCASEE24 pKa = 3.88EE25 pKa = 4.0AGLVGEE31 pKa = 4.08WHH33 pKa = 6.85FNEE36 pKa = 4.03TDD38 pKa = 3.15GNIAYY43 pKa = 9.64DD44 pKa = 3.87FSGNGNDD51 pKa = 3.74GTINGASWVGGVSGSALSFDD71 pKa = 3.66GNDD74 pKa = 3.47YY75 pKa = 11.57VNTNLHH81 pKa = 6.53LPQGSKK87 pKa = 8.3TVEE90 pKa = 3.26AWIKK94 pKa = 11.01GFGTTYY100 pKa = 9.75STEE103 pKa = 3.97CPIAGEE109 pKa = 4.12RR110 pKa = 11.84WYY112 pKa = 10.88SAAGGGEE119 pKa = 3.84QLNLGVKK126 pKa = 8.71TDD128 pKa = 3.94EE129 pKa = 4.17SAMLYY134 pKa = 10.29VYY136 pKa = 10.67SEE138 pKa = 4.57DD139 pKa = 3.6KK140 pKa = 11.16GSGVNAVSDD149 pKa = 4.67SNILDD154 pKa = 3.36QTKK157 pKa = 7.77WHH159 pKa = 7.06HH160 pKa = 5.83IAGTYY165 pKa = 9.21EE166 pKa = 3.95VGDD169 pKa = 3.6TDD171 pKa = 4.51AVLKK175 pKa = 10.49IYY177 pKa = 10.32IDD179 pKa = 3.77GTLHH183 pKa = 6.27DD184 pKa = 4.65TVHH187 pKa = 6.5GPIDD191 pKa = 3.78IDD193 pKa = 3.67NHH195 pKa = 5.4ISTYY199 pKa = 10.7LIGVWRR205 pKa = 11.84RR206 pKa = 11.84GTGAVGVRR214 pKa = 11.84YY215 pKa = 9.22FKK217 pKa = 11.17GVIDD221 pKa = 3.79EE222 pKa = 4.0VRR224 pKa = 11.84IYY226 pKa = 11.17NRR228 pKa = 11.84ALTATEE234 pKa = 3.53ISDD237 pKa = 4.47RR238 pKa = 11.84YY239 pKa = 8.55YY240 pKa = 10.32QQDD243 pKa = 3.29MDD245 pKa = 4.45RR246 pKa = 11.84DD247 pKa = 3.85GVLNINDD254 pKa = 3.54QCPNTTAKK262 pKa = 10.82CEE264 pKa = 4.07VDD266 pKa = 4.28SSGCPLDD273 pKa = 4.05SDD275 pKa = 5.01NDD277 pKa = 4.15NICNGIDD284 pKa = 3.45TCPNTPLNCNVSSTGCKK301 pKa = 8.95IDD303 pKa = 3.53SDD305 pKa = 4.43SDD307 pKa = 4.33GVCNGVDD314 pKa = 3.47PCPNDD319 pKa = 5.64LEE321 pKa = 4.66DD322 pKa = 4.48CCTATTDD329 pKa = 3.54TDD331 pKa = 3.53NDD333 pKa = 3.95SLQEE337 pKa = 4.27SNSNCPNYY345 pKa = 10.46DD346 pKa = 3.6SYY348 pKa = 12.06PNDD351 pKa = 3.56HH352 pKa = 7.51DD353 pKa = 3.99NDD355 pKa = 4.31GVVSSVDD362 pKa = 4.0CDD364 pKa = 4.46DD365 pKa = 4.6SDD367 pKa = 5.0AGNTKK372 pKa = 9.58TKK374 pKa = 10.67GDD376 pKa = 3.91SDD378 pKa = 3.98NDD380 pKa = 4.06GVDD383 pKa = 3.34NCIDD387 pKa = 3.68SCPNDD392 pKa = 3.64AGNQCGCVDD401 pKa = 3.97HH402 pKa = 7.47DD403 pKa = 4.73GDD405 pKa = 4.84GLGEE409 pKa = 4.19GAGCRR414 pKa = 11.84DD415 pKa = 3.66PCSNDD420 pKa = 3.27KK421 pKa = 11.11DD422 pKa = 4.09NCCTAITDD430 pKa = 3.71TDD432 pKa = 3.62NDD434 pKa = 3.92GLQEE438 pKa = 4.28NNVNCPNYY446 pKa = 10.6DD447 pKa = 3.35NYY449 pKa = 11.47LNDD452 pKa = 3.84HH453 pKa = 7.27DD454 pKa = 4.59NDD456 pKa = 4.26GVISSVDD463 pKa = 3.78CDD465 pKa = 5.12DD466 pKa = 4.71SDD468 pKa = 5.0AGNTKK473 pKa = 9.56TKK475 pKa = 10.56GDD477 pKa = 3.81SDD479 pKa = 4.19GDD481 pKa = 4.01GLNNCQDD488 pKa = 3.67SCPDD492 pKa = 4.03DD493 pKa = 5.42ANNQCGCVDD502 pKa = 3.53NDD504 pKa = 3.78GDD506 pKa = 4.38GLGEE510 pKa = 4.19GTGCADD516 pKa = 3.83PCPNDD521 pKa = 4.79RR522 pKa = 11.84DD523 pKa = 3.95NCCIATTDD531 pKa = 3.76ADD533 pKa = 3.77NDD535 pKa = 3.55NLQEE539 pKa = 4.27NNQDD543 pKa = 3.41CPNYY547 pKa = 10.58DD548 pKa = 3.55NYY550 pKa = 11.48LNDD553 pKa = 3.88HH554 pKa = 7.24DD555 pKa = 4.82NDD557 pKa = 4.19GVTTLMDD564 pKa = 5.18CDD566 pKa = 5.76DD567 pKa = 4.41SDD569 pKa = 4.72ADD571 pKa = 3.73NVKK574 pKa = 10.34IKK576 pKa = 10.62GDD578 pKa = 4.08CGDD581 pKa = 3.5QEE583 pKa = 4.53QIPVVNVTVVNEE595 pKa = 3.73ITVINTITIANEE607 pKa = 4.3TIFSAVTTINEE618 pKa = 3.73TRR620 pKa = 11.84RR621 pKa = 11.84YY622 pKa = 9.63LNVSEE627 pKa = 4.21EE628 pKa = 4.27VNLEE632 pKa = 3.85VEE634 pKa = 4.53EE635 pKa = 4.57KK636 pKa = 10.23IKK638 pKa = 10.65EE639 pKa = 4.07VEE641 pKa = 4.16DD642 pKa = 4.13LIEE645 pKa = 4.07RR646 pKa = 11.84AKK648 pKa = 10.98EE649 pKa = 3.87KK650 pKa = 10.47IALANSLINTDD661 pKa = 3.74PKK663 pKa = 10.82RR664 pKa = 11.84AMDD667 pKa = 3.58EE668 pKa = 4.16AEE670 pKa = 4.37EE671 pKa = 4.28SNEE674 pKa = 3.73LAEE677 pKa = 4.32RR678 pKa = 11.84AVKK681 pKa = 9.07LTMEE685 pKa = 4.52LSKK688 pKa = 11.04NYY690 pKa = 7.99KK691 pKa = 8.64TKK693 pKa = 10.91SSVLPATSQSDD704 pKa = 3.54ALVSEE709 pKa = 4.62TEE711 pKa = 4.24EE712 pKa = 4.23FSGDD716 pKa = 3.25TFKK719 pKa = 9.31WTGIAGIVCFIILTGILIFAARR741 pKa = 11.84KK742 pKa = 9.59KK743 pKa = 10.24YY744 pKa = 10.59
MM1 pKa = 6.85KK2 pKa = 9.63TSNIILLIFVFEE14 pKa = 4.97IILSNLCASEE24 pKa = 3.88EE25 pKa = 4.0AGLVGEE31 pKa = 4.08WHH33 pKa = 6.85FNEE36 pKa = 4.03TDD38 pKa = 3.15GNIAYY43 pKa = 9.64DD44 pKa = 3.87FSGNGNDD51 pKa = 3.74GTINGASWVGGVSGSALSFDD71 pKa = 3.66GNDD74 pKa = 3.47YY75 pKa = 11.57VNTNLHH81 pKa = 6.53LPQGSKK87 pKa = 8.3TVEE90 pKa = 3.26AWIKK94 pKa = 11.01GFGTTYY100 pKa = 9.75STEE103 pKa = 3.97CPIAGEE109 pKa = 4.12RR110 pKa = 11.84WYY112 pKa = 10.88SAAGGGEE119 pKa = 3.84QLNLGVKK126 pKa = 8.71TDD128 pKa = 3.94EE129 pKa = 4.17SAMLYY134 pKa = 10.29VYY136 pKa = 10.67SEE138 pKa = 4.57DD139 pKa = 3.6KK140 pKa = 11.16GSGVNAVSDD149 pKa = 4.67SNILDD154 pKa = 3.36QTKK157 pKa = 7.77WHH159 pKa = 7.06HH160 pKa = 5.83IAGTYY165 pKa = 9.21EE166 pKa = 3.95VGDD169 pKa = 3.6TDD171 pKa = 4.51AVLKK175 pKa = 10.49IYY177 pKa = 10.32IDD179 pKa = 3.77GTLHH183 pKa = 6.27DD184 pKa = 4.65TVHH187 pKa = 6.5GPIDD191 pKa = 3.78IDD193 pKa = 3.67NHH195 pKa = 5.4ISTYY199 pKa = 10.7LIGVWRR205 pKa = 11.84RR206 pKa = 11.84GTGAVGVRR214 pKa = 11.84YY215 pKa = 9.22FKK217 pKa = 11.17GVIDD221 pKa = 3.79EE222 pKa = 4.0VRR224 pKa = 11.84IYY226 pKa = 11.17NRR228 pKa = 11.84ALTATEE234 pKa = 3.53ISDD237 pKa = 4.47RR238 pKa = 11.84YY239 pKa = 8.55YY240 pKa = 10.32QQDD243 pKa = 3.29MDD245 pKa = 4.45RR246 pKa = 11.84DD247 pKa = 3.85GVLNINDD254 pKa = 3.54QCPNTTAKK262 pKa = 10.82CEE264 pKa = 4.07VDD266 pKa = 4.28SSGCPLDD273 pKa = 4.05SDD275 pKa = 5.01NDD277 pKa = 4.15NICNGIDD284 pKa = 3.45TCPNTPLNCNVSSTGCKK301 pKa = 8.95IDD303 pKa = 3.53SDD305 pKa = 4.43SDD307 pKa = 4.33GVCNGVDD314 pKa = 3.47PCPNDD319 pKa = 5.64LEE321 pKa = 4.66DD322 pKa = 4.48CCTATTDD329 pKa = 3.54TDD331 pKa = 3.53NDD333 pKa = 3.95SLQEE337 pKa = 4.27SNSNCPNYY345 pKa = 10.46DD346 pKa = 3.6SYY348 pKa = 12.06PNDD351 pKa = 3.56HH352 pKa = 7.51DD353 pKa = 3.99NDD355 pKa = 4.31GVVSSVDD362 pKa = 4.0CDD364 pKa = 4.46DD365 pKa = 4.6SDD367 pKa = 5.0AGNTKK372 pKa = 9.58TKK374 pKa = 10.67GDD376 pKa = 3.91SDD378 pKa = 3.98NDD380 pKa = 4.06GVDD383 pKa = 3.34NCIDD387 pKa = 3.68SCPNDD392 pKa = 3.64AGNQCGCVDD401 pKa = 3.97HH402 pKa = 7.47DD403 pKa = 4.73GDD405 pKa = 4.84GLGEE409 pKa = 4.19GAGCRR414 pKa = 11.84DD415 pKa = 3.66PCSNDD420 pKa = 3.27KK421 pKa = 11.11DD422 pKa = 4.09NCCTAITDD430 pKa = 3.71TDD432 pKa = 3.62NDD434 pKa = 3.92GLQEE438 pKa = 4.28NNVNCPNYY446 pKa = 10.6DD447 pKa = 3.35NYY449 pKa = 11.47LNDD452 pKa = 3.84HH453 pKa = 7.27DD454 pKa = 4.59NDD456 pKa = 4.26GVISSVDD463 pKa = 3.78CDD465 pKa = 5.12DD466 pKa = 4.71SDD468 pKa = 5.0AGNTKK473 pKa = 9.56TKK475 pKa = 10.56GDD477 pKa = 3.81SDD479 pKa = 4.19GDD481 pKa = 4.01GLNNCQDD488 pKa = 3.67SCPDD492 pKa = 4.03DD493 pKa = 5.42ANNQCGCVDD502 pKa = 3.53NDD504 pKa = 3.78GDD506 pKa = 4.38GLGEE510 pKa = 4.19GTGCADD516 pKa = 3.83PCPNDD521 pKa = 4.79RR522 pKa = 11.84DD523 pKa = 3.95NCCIATTDD531 pKa = 3.76ADD533 pKa = 3.77NDD535 pKa = 3.55NLQEE539 pKa = 4.27NNQDD543 pKa = 3.41CPNYY547 pKa = 10.58DD548 pKa = 3.55NYY550 pKa = 11.48LNDD553 pKa = 3.88HH554 pKa = 7.24DD555 pKa = 4.82NDD557 pKa = 4.19GVTTLMDD564 pKa = 5.18CDD566 pKa = 5.76DD567 pKa = 4.41SDD569 pKa = 4.72ADD571 pKa = 3.73NVKK574 pKa = 10.34IKK576 pKa = 10.62GDD578 pKa = 4.08CGDD581 pKa = 3.5QEE583 pKa = 4.53QIPVVNVTVVNEE595 pKa = 3.73ITVINTITIANEE607 pKa = 4.3TIFSAVTTINEE618 pKa = 3.73TRR620 pKa = 11.84RR621 pKa = 11.84YY622 pKa = 9.63LNVSEE627 pKa = 4.21EE628 pKa = 4.27VNLEE632 pKa = 3.85VEE634 pKa = 4.53EE635 pKa = 4.57KK636 pKa = 10.23IKK638 pKa = 10.65EE639 pKa = 4.07VEE641 pKa = 4.16DD642 pKa = 4.13LIEE645 pKa = 4.07RR646 pKa = 11.84AKK648 pKa = 10.98EE649 pKa = 3.87KK650 pKa = 10.47IALANSLINTDD661 pKa = 3.74PKK663 pKa = 10.82RR664 pKa = 11.84AMDD667 pKa = 3.58EE668 pKa = 4.16AEE670 pKa = 4.37EE671 pKa = 4.28SNEE674 pKa = 3.73LAEE677 pKa = 4.32RR678 pKa = 11.84AVKK681 pKa = 9.07LTMEE685 pKa = 4.52LSKK688 pKa = 11.04NYY690 pKa = 7.99KK691 pKa = 8.64TKK693 pKa = 10.91SSVLPATSQSDD704 pKa = 3.54ALVSEE709 pKa = 4.62TEE711 pKa = 4.24EE712 pKa = 4.23FSGDD716 pKa = 3.25TFKK719 pKa = 9.31WTGIAGIVCFIILTGILIFAARR741 pKa = 11.84KK742 pKa = 9.59KK743 pKa = 10.24YY744 pKa = 10.59
Molecular weight: 80.03 kDa
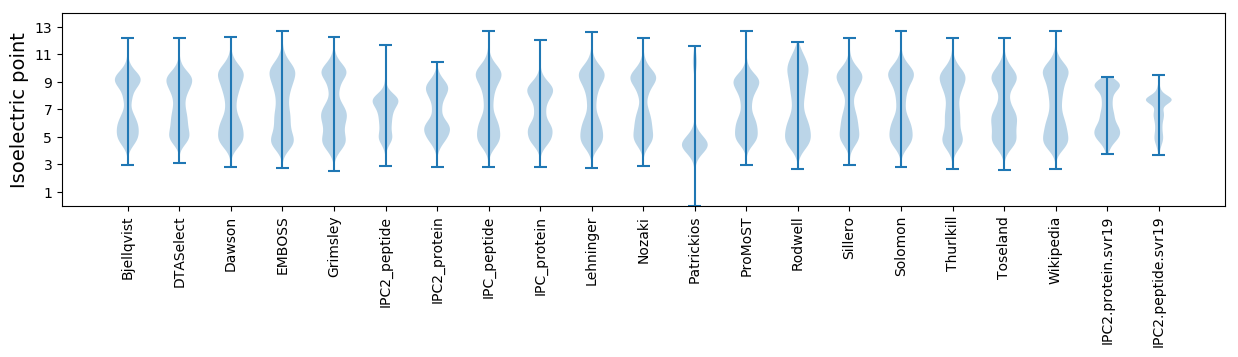
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1D2QUD6|A0A1D2QUD6_9ARCH PQQ_3 domain-containing protein OS=Candidatus Altiarchaeales archaeon WOR_SM1_SCG OX=1849261 GN=BEH94_01960 PE=4 SV=1
MM1 pKa = 7.26YY2 pKa = 10.79SHH4 pKa = 6.87TKK6 pKa = 9.9KK7 pKa = 10.34VGSIGRR13 pKa = 11.84YY14 pKa = 7.07GPRR17 pKa = 11.84VGRR20 pKa = 11.84KK21 pKa = 7.36VRR23 pKa = 11.84MAMRR27 pKa = 11.84KK28 pKa = 9.03IEE30 pKa = 4.51DD31 pKa = 3.46KK32 pKa = 10.64TRR34 pKa = 11.84EE35 pKa = 4.0ANKK38 pKa = 10.17CKK40 pKa = 10.62SCGKK44 pKa = 9.26PKK46 pKa = 9.9IRR48 pKa = 11.84RR49 pKa = 11.84EE50 pKa = 3.77SAGIWKK56 pKa = 9.46CRR58 pKa = 11.84ACGVTFTGGAHH69 pKa = 6.47LPVTEE74 pKa = 4.27RR75 pKa = 11.84RR76 pKa = 3.73
MM1 pKa = 7.26YY2 pKa = 10.79SHH4 pKa = 6.87TKK6 pKa = 9.9KK7 pKa = 10.34VGSIGRR13 pKa = 11.84YY14 pKa = 7.07GPRR17 pKa = 11.84VGRR20 pKa = 11.84KK21 pKa = 7.36VRR23 pKa = 11.84MAMRR27 pKa = 11.84KK28 pKa = 9.03IEE30 pKa = 4.51DD31 pKa = 3.46KK32 pKa = 10.64TRR34 pKa = 11.84EE35 pKa = 4.0ANKK38 pKa = 10.17CKK40 pKa = 10.62SCGKK44 pKa = 9.26PKK46 pKa = 9.9IRR48 pKa = 11.84RR49 pKa = 11.84EE50 pKa = 3.77SAGIWKK56 pKa = 9.46CRR58 pKa = 11.84ACGVTFTGGAHH69 pKa = 6.47LPVTEE74 pKa = 4.27RR75 pKa = 11.84RR76 pKa = 3.73
Molecular weight: 8.58 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
656850 |
48 |
5438 |
303.5 |
34.24 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.755 ± 0.058 | 1.631 ± 0.052 |
5.798 ± 0.053 | 7.891 ± 0.071 |
4.31 ± 0.049 | 6.904 ± 0.052 |
1.629 ± 0.022 | 9.204 ± 0.054 |
8.965 ± 0.103 | 8.459 ± 0.073 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.274 ± 0.028 | 5.937 ± 0.115 |
3.401 ± 0.034 | 2.024 ± 0.026 |
3.914 ± 0.049 | 5.869 ± 0.057 |
5.085 ± 0.082 | 6.252 ± 0.049 |
0.958 ± 0.02 | 3.738 ± 0.053 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
