
Marinospirillum celere
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Oceanospirillales; Oceanospirillaceae; Marinospirillum
Average proteome isoelectric point is 6.0
Get precalculated fractions of proteins
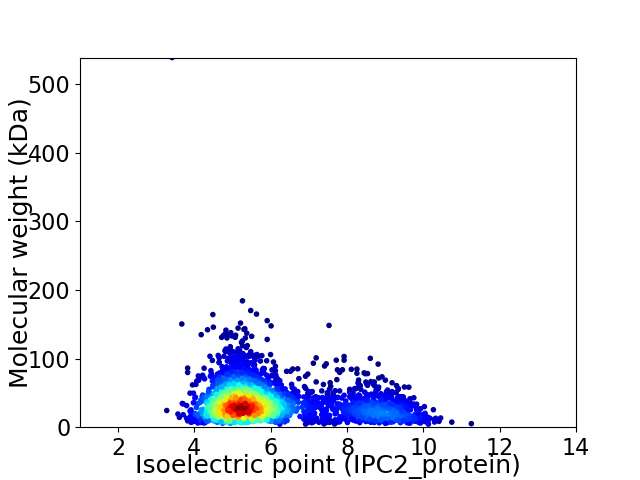
Virtual 2D-PAGE plot for 2818 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
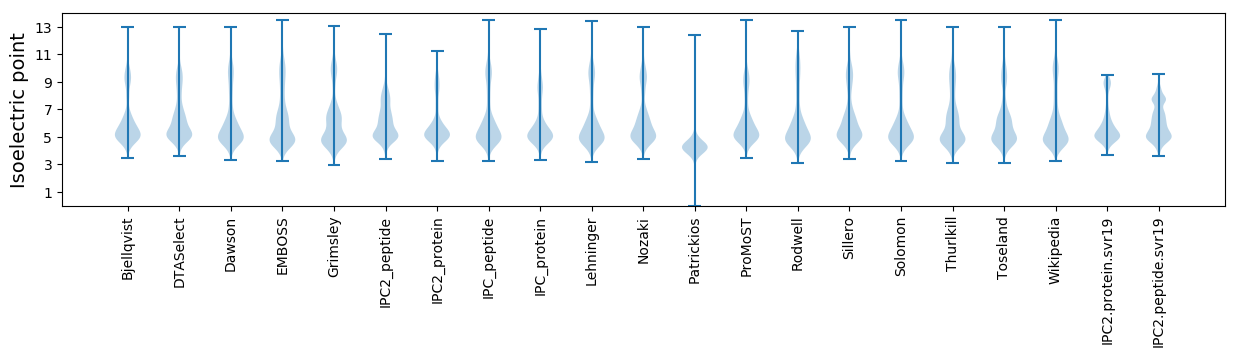
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I1G398|A0A1I1G398_9GAMM Ribosome-associated protein OS=Marinospirillum celere OX=1122252 GN=SAMN05660443_1256 PE=4 SV=1
MM1 pKa = 7.57KK2 pKa = 10.38KK3 pKa = 10.61LNLALLISLGWLLTACLAEE22 pKa = 3.96QDD24 pKa = 3.6EE25 pKa = 4.86SSRR28 pKa = 11.84QLGSDD33 pKa = 3.43ASQGGEE39 pKa = 3.79YY40 pKa = 10.22RR41 pKa = 11.84QGEE44 pKa = 4.11GTARR48 pKa = 11.84VEE50 pKa = 4.58FSVHH54 pKa = 5.82LPGDD58 pKa = 3.57ASAAYY63 pKa = 9.35IDD65 pKa = 4.04TEE67 pKa = 4.16NTRR70 pKa = 11.84SIVVSVFPSSIAGFDD85 pKa = 3.28VDD87 pKa = 5.06RR88 pKa = 11.84FQATDD93 pKa = 3.05VLRR96 pKa = 11.84AYY98 pKa = 7.3FQCVMANAPSEE109 pKa = 3.99GDD111 pKa = 3.17MGIQEE116 pKa = 4.45MEE118 pKa = 4.58SACGRR123 pKa = 11.84YY124 pKa = 8.15PAQIADD130 pKa = 3.92TPLYY134 pKa = 10.61SATLTSDD141 pKa = 3.93LPAAGFNLQPGNSYY155 pKa = 10.34IVLAAQYY162 pKa = 10.5SSSNPAGLTPVGWTTTTITPNDD184 pKa = 3.67GNNDD188 pKa = 3.2VEE190 pKa = 5.28LNLLYY195 pKa = 10.72GSWEE199 pKa = 3.94FDD201 pKa = 3.12GGPSLQLLNRR211 pKa = 11.84STYY214 pKa = 11.31AFGNFIGFNEE224 pKa = 3.93EE225 pKa = 3.8DD226 pKa = 3.45LEE228 pKa = 4.49EE229 pKa = 4.2FTEE232 pKa = 5.59APDD235 pKa = 2.86WWPASTHH242 pKa = 5.69VMSTPAMALGLADD255 pKa = 4.23TGDD258 pKa = 3.34ATINLDD264 pKa = 3.67RR265 pKa = 11.84LHH267 pKa = 6.51LRR269 pKa = 11.84SYY271 pKa = 11.62YY272 pKa = 10.14EE273 pKa = 4.0LSPNNQVDD281 pKa = 3.65SARR284 pKa = 11.84NDD286 pKa = 3.03EE287 pKa = 4.52GIRR290 pKa = 11.84LFGAQGEE297 pKa = 4.37PDD299 pKa = 3.55NFDD302 pKa = 5.13LFGQYY307 pKa = 9.43PWGILAPHH315 pKa = 6.76EE316 pKa = 4.69PLLQSSVAGNAFFVEE331 pKa = 4.38PRR333 pKa = 11.84FEE335 pKa = 4.13LVEE338 pKa = 5.09GIDD341 pKa = 4.74DD342 pKa = 5.65DD343 pKa = 4.78PDD345 pKa = 4.25LMLGRR350 pKa = 11.84GVYY353 pKa = 10.01EE354 pKa = 4.57PATLMQQYY362 pKa = 10.66APEE365 pKa = 3.73QSQANRR371 pKa = 11.84HH372 pKa = 5.13RR373 pKa = 11.84LGLGTWEE380 pKa = 3.96VTIGEE385 pKa = 4.52EE386 pKa = 3.74ISTEE390 pKa = 3.93NVEE393 pKa = 4.07IFAGLMLVSGEE404 pKa = 3.92NDD406 pKa = 2.9ILNLLRR412 pKa = 11.84DD413 pKa = 3.58ATEE416 pKa = 3.77EE417 pKa = 4.25VEE419 pKa = 4.41GQRR422 pKa = 11.84SFDD425 pKa = 3.43QLNVRR430 pKa = 11.84RR431 pKa = 11.84PVFDD435 pKa = 3.3QFGQFGQFGQFGQFGQFGQPVPEE458 pKa = 4.28VLPEE462 pKa = 4.26SNLTIAFFDD471 pKa = 3.86IEE473 pKa = 4.47TQQGEE478 pKa = 4.35LLIAEE483 pKa = 4.64GTDD486 pKa = 3.36TQNFQALDD494 pKa = 3.47ALEE497 pKa = 4.21PTRR500 pKa = 11.84LTAADD505 pKa = 5.37RR506 pKa = 11.84IQGSLIEE513 pKa = 4.13YY514 pKa = 8.41VGYY517 pKa = 10.87YY518 pKa = 9.83NAIEE522 pKa = 4.35SPPYY526 pKa = 10.08QYY528 pKa = 11.22PLLLEE533 pKa = 4.79DD534 pKa = 3.98EE535 pKa = 5.02EE536 pKa = 6.06GNFTQVDD543 pKa = 4.42TIEE546 pKa = 4.14VNPVIGGVAASSSLQNTSGSSLRR569 pKa = 11.84SSSQALILRR578 pKa = 11.84NLQLSALAADD588 pKa = 4.11SGLIASTAASGCFVMDD604 pKa = 4.91LDD606 pKa = 4.13VQRR609 pKa = 11.84VSSFYY614 pKa = 10.93LFNPDD619 pKa = 3.17EE620 pKa = 4.47EE621 pKa = 4.79TWLPAAPEE629 pKa = 3.79FGYY632 pKa = 9.74WYY634 pKa = 9.74QAEE637 pKa = 4.45DD638 pKa = 3.4GSYY641 pKa = 11.01LRR643 pKa = 11.84DD644 pKa = 3.41EE645 pKa = 4.46EE646 pKa = 4.97GKK648 pKa = 9.9IEE650 pKa = 4.16TFGTGASPSEE660 pKa = 4.0TLEE663 pKa = 3.98EE664 pKa = 4.63DD665 pKa = 3.65GVIYY669 pKa = 10.28SIQPSGGMTGIIDD682 pKa = 4.15ASTDD686 pKa = 3.53EE687 pKa = 5.34SDD689 pKa = 4.81LEE691 pKa = 4.23QNFYY695 pKa = 10.95AWSEE699 pKa = 4.04EE700 pKa = 3.92GSGKK704 pKa = 10.08VEE706 pKa = 4.01VCIQPVSLTAGPYY719 pKa = 10.5SNPEE723 pKa = 3.77YY724 pKa = 10.35NAEE727 pKa = 3.97SDD729 pKa = 4.4VVIQQ733 pKa = 4.44
MM1 pKa = 7.57KK2 pKa = 10.38KK3 pKa = 10.61LNLALLISLGWLLTACLAEE22 pKa = 3.96QDD24 pKa = 3.6EE25 pKa = 4.86SSRR28 pKa = 11.84QLGSDD33 pKa = 3.43ASQGGEE39 pKa = 3.79YY40 pKa = 10.22RR41 pKa = 11.84QGEE44 pKa = 4.11GTARR48 pKa = 11.84VEE50 pKa = 4.58FSVHH54 pKa = 5.82LPGDD58 pKa = 3.57ASAAYY63 pKa = 9.35IDD65 pKa = 4.04TEE67 pKa = 4.16NTRR70 pKa = 11.84SIVVSVFPSSIAGFDD85 pKa = 3.28VDD87 pKa = 5.06RR88 pKa = 11.84FQATDD93 pKa = 3.05VLRR96 pKa = 11.84AYY98 pKa = 7.3FQCVMANAPSEE109 pKa = 3.99GDD111 pKa = 3.17MGIQEE116 pKa = 4.45MEE118 pKa = 4.58SACGRR123 pKa = 11.84YY124 pKa = 8.15PAQIADD130 pKa = 3.92TPLYY134 pKa = 10.61SATLTSDD141 pKa = 3.93LPAAGFNLQPGNSYY155 pKa = 10.34IVLAAQYY162 pKa = 10.5SSSNPAGLTPVGWTTTTITPNDD184 pKa = 3.67GNNDD188 pKa = 3.2VEE190 pKa = 5.28LNLLYY195 pKa = 10.72GSWEE199 pKa = 3.94FDD201 pKa = 3.12GGPSLQLLNRR211 pKa = 11.84STYY214 pKa = 11.31AFGNFIGFNEE224 pKa = 3.93EE225 pKa = 3.8DD226 pKa = 3.45LEE228 pKa = 4.49EE229 pKa = 4.2FTEE232 pKa = 5.59APDD235 pKa = 2.86WWPASTHH242 pKa = 5.69VMSTPAMALGLADD255 pKa = 4.23TGDD258 pKa = 3.34ATINLDD264 pKa = 3.67RR265 pKa = 11.84LHH267 pKa = 6.51LRR269 pKa = 11.84SYY271 pKa = 11.62YY272 pKa = 10.14EE273 pKa = 4.0LSPNNQVDD281 pKa = 3.65SARR284 pKa = 11.84NDD286 pKa = 3.03EE287 pKa = 4.52GIRR290 pKa = 11.84LFGAQGEE297 pKa = 4.37PDD299 pKa = 3.55NFDD302 pKa = 5.13LFGQYY307 pKa = 9.43PWGILAPHH315 pKa = 6.76EE316 pKa = 4.69PLLQSSVAGNAFFVEE331 pKa = 4.38PRR333 pKa = 11.84FEE335 pKa = 4.13LVEE338 pKa = 5.09GIDD341 pKa = 4.74DD342 pKa = 5.65DD343 pKa = 4.78PDD345 pKa = 4.25LMLGRR350 pKa = 11.84GVYY353 pKa = 10.01EE354 pKa = 4.57PATLMQQYY362 pKa = 10.66APEE365 pKa = 3.73QSQANRR371 pKa = 11.84HH372 pKa = 5.13RR373 pKa = 11.84LGLGTWEE380 pKa = 3.96VTIGEE385 pKa = 4.52EE386 pKa = 3.74ISTEE390 pKa = 3.93NVEE393 pKa = 4.07IFAGLMLVSGEE404 pKa = 3.92NDD406 pKa = 2.9ILNLLRR412 pKa = 11.84DD413 pKa = 3.58ATEE416 pKa = 3.77EE417 pKa = 4.25VEE419 pKa = 4.41GQRR422 pKa = 11.84SFDD425 pKa = 3.43QLNVRR430 pKa = 11.84RR431 pKa = 11.84PVFDD435 pKa = 3.3QFGQFGQFGQFGQFGQFGQPVPEE458 pKa = 4.28VLPEE462 pKa = 4.26SNLTIAFFDD471 pKa = 3.86IEE473 pKa = 4.47TQQGEE478 pKa = 4.35LLIAEE483 pKa = 4.64GTDD486 pKa = 3.36TQNFQALDD494 pKa = 3.47ALEE497 pKa = 4.21PTRR500 pKa = 11.84LTAADD505 pKa = 5.37RR506 pKa = 11.84IQGSLIEE513 pKa = 4.13YY514 pKa = 8.41VGYY517 pKa = 10.87YY518 pKa = 9.83NAIEE522 pKa = 4.35SPPYY526 pKa = 10.08QYY528 pKa = 11.22PLLLEE533 pKa = 4.79DD534 pKa = 3.98EE535 pKa = 5.02EE536 pKa = 6.06GNFTQVDD543 pKa = 4.42TIEE546 pKa = 4.14VNPVIGGVAASSSLQNTSGSSLRR569 pKa = 11.84SSSQALILRR578 pKa = 11.84NLQLSALAADD588 pKa = 4.11SGLIASTAASGCFVMDD604 pKa = 4.91LDD606 pKa = 4.13VQRR609 pKa = 11.84VSSFYY614 pKa = 10.93LFNPDD619 pKa = 3.17EE620 pKa = 4.47EE621 pKa = 4.79TWLPAAPEE629 pKa = 3.79FGYY632 pKa = 9.74WYY634 pKa = 9.74QAEE637 pKa = 4.45DD638 pKa = 3.4GSYY641 pKa = 11.01LRR643 pKa = 11.84DD644 pKa = 3.41EE645 pKa = 4.46EE646 pKa = 4.97GKK648 pKa = 9.9IEE650 pKa = 4.16TFGTGASPSEE660 pKa = 4.0TLEE663 pKa = 3.98EE664 pKa = 4.63DD665 pKa = 3.65GVIYY669 pKa = 10.28SIQPSGGMTGIIDD682 pKa = 4.15ASTDD686 pKa = 3.53EE687 pKa = 5.34SDD689 pKa = 4.81LEE691 pKa = 4.23QNFYY695 pKa = 10.95AWSEE699 pKa = 4.04EE700 pKa = 3.92GSGKK704 pKa = 10.08VEE706 pKa = 4.01VCIQPVSLTAGPYY719 pKa = 10.5SNPEE723 pKa = 3.77YY724 pKa = 10.35NAEE727 pKa = 3.97SDD729 pKa = 4.4VVIQQ733 pKa = 4.44
Molecular weight: 79.81 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I1IAE0|A0A1I1IAE0_9GAMM Sulfur carrier protein OS=Marinospirillum celere OX=1122252 GN=SAMN05660443_2299 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.46RR12 pKa = 11.84KK13 pKa = 9.37RR14 pKa = 11.84NHH16 pKa = 5.37GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.3NGRR28 pKa = 11.84AVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.62GRR39 pKa = 11.84KK40 pKa = 9.11RR41 pKa = 11.84ISAA44 pKa = 3.72
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.46RR12 pKa = 11.84KK13 pKa = 9.37RR14 pKa = 11.84NHH16 pKa = 5.37GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.3NGRR28 pKa = 11.84AVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.62GRR39 pKa = 11.84KK40 pKa = 9.11RR41 pKa = 11.84ISAA44 pKa = 3.72
Molecular weight: 5.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
946962 |
37 |
5359 |
336.0 |
37.35 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.535 ± 0.051 | 0.87 ± 0.015 |
5.301 ± 0.039 | 7.06 ± 0.051 |
3.728 ± 0.031 | 7.125 ± 0.058 |
2.163 ± 0.021 | 4.912 ± 0.039 |
4.225 ± 0.046 | 12.509 ± 0.077 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.279 ± 0.026 | 3.255 ± 0.04 |
4.533 ± 0.04 | 5.63 ± 0.056 |
5.757 ± 0.052 | 5.86 ± 0.039 |
4.874 ± 0.07 | 6.363 ± 0.04 |
1.467 ± 0.024 | 2.553 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
