
Methylobacterium sp. 17Sr1-43
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Methylobacteriaceae; Methylobacterium
Average proteome isoelectric point is 6.91
Get precalculated fractions of proteins
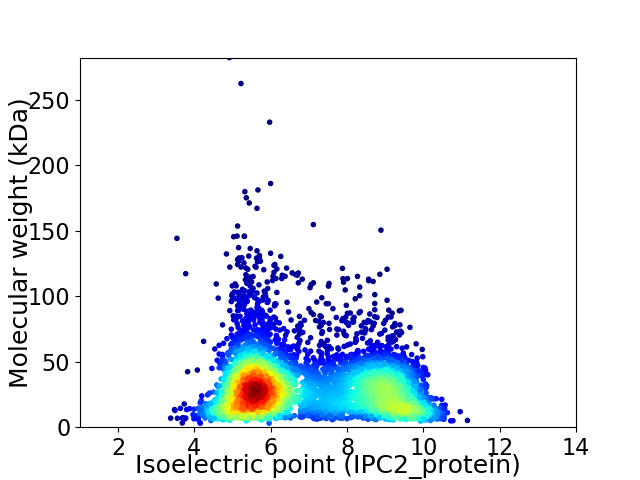
Virtual 2D-PAGE plot for 4920 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2U8VYZ9|A0A2U8VYZ9_9RHIZ Putative membrane protein insertion efficiency factor OS=Methylobacterium sp. 17Sr1-43 OX=2202828 GN=DK427_14665 PE=3 SV=1
MM1 pKa = 7.79PATFGTATTTLVEE14 pKa = 4.24SEE16 pKa = 4.03PWEE19 pKa = 4.45LSAADD24 pKa = 4.13LNGDD28 pKa = 3.67GKK30 pKa = 11.33LDD32 pKa = 3.47LVVPNTASNSLSVLIGNGDD51 pKa = 3.52GSFRR55 pKa = 11.84PAVNYY60 pKa = 8.21PVSSSPISGALADD73 pKa = 3.65VTGDD77 pKa = 3.47GKK79 pKa = 10.64IDD81 pKa = 3.29IVVGINGAPSLLAGNGDD98 pKa = 3.84GTFQAAAGIAGGNALQSYY116 pKa = 7.12WLRR119 pKa = 11.84AVDD122 pKa = 4.87LNADD126 pKa = 3.79GKK128 pKa = 11.42LDD130 pKa = 3.79LAYY133 pKa = 9.96TNFGPNQLVVQYY145 pKa = 10.94GNGDD149 pKa = 3.6GTFGTGALYY158 pKa = 10.66SVGNLPRR165 pKa = 11.84APAIGDD171 pKa = 3.65FNHH174 pKa = 7.23DD175 pKa = 3.52GKK177 pKa = 11.43LDD179 pKa = 3.41IVTANSGATSVSVLLANADD198 pKa = 3.33GTYY201 pKa = 10.24QSQVPYY207 pKa = 9.99PVGASPYY214 pKa = 10.39SVATGDD220 pKa = 3.89FNGDD224 pKa = 3.14GNLDD228 pKa = 3.77LVTAALNANSYY239 pKa = 10.94SVLLGRR245 pKa = 11.84GDD247 pKa = 3.8GTFQPALTTAAQGNSNEE264 pKa = 4.06ITVADD269 pKa = 3.54VDD271 pKa = 4.22GDD273 pKa = 4.03GKK275 pKa = 11.2LDD277 pKa = 4.58LIIANSSANQVGIAYY292 pKa = 8.89GQGDD296 pKa = 4.18GTFSAWQTLPTGGGPYY312 pKa = 9.1TAIAVDD318 pKa = 4.05LNGDD322 pKa = 3.41GRR324 pKa = 11.84LDD326 pKa = 3.54LAVTNRR332 pKa = 11.84VSGTVTTYY340 pKa = 8.83MQSTPPTLSITGATVVEE357 pKa = 4.96GNSGTADD364 pKa = 3.11AAFVVTRR371 pKa = 11.84SDD373 pKa = 3.48TTGSPTATYY382 pKa = 9.2TITNSTTDD390 pKa = 3.35GADD393 pKa = 3.5FGPGFQATGTVTFAAGATTATINVPIRR420 pKa = 11.84GDD422 pKa = 3.52TLYY425 pKa = 10.57EE426 pKa = 3.92ASEE429 pKa = 4.49TFTVALSNPQGATISGTGQATGTITNDD456 pKa = 3.36DD457 pKa = 4.11APPTVTLSGGTSIAEE472 pKa = 4.42GNSGTTVVPITATLSAVSGLATTVTLAFGGTATQGTDD509 pKa = 3.32YY510 pKa = 9.81TVSSTTITIPAGSLSGSVNVNVLGDD535 pKa = 3.65TAVEE539 pKa = 4.16PNGTATVTIATATNATPAGAAQTVTIQNDD568 pKa = 3.65DD569 pKa = 3.0VSYY572 pKa = 11.17AITASAAAVAEE583 pKa = 4.65GSSGAGNAVTYY594 pKa = 9.13TVTRR598 pKa = 11.84TGDD601 pKa = 3.17MTQAGVATVRR611 pKa = 11.84LSGEE615 pKa = 3.81ATRR618 pKa = 11.84GDD620 pKa = 4.46DD621 pKa = 4.38YY622 pKa = 11.55STTLPANGQINFVAGDD638 pKa = 3.53ASKK641 pKa = 9.48TFTVTTVPDD650 pKa = 3.57TTVEE654 pKa = 3.91PNEE657 pKa = 4.07RR658 pKa = 11.84VIATITGVSGQGGATIGTPASATTTITNDD687 pKa = 3.13DD688 pKa = 3.75QYY690 pKa = 11.49GVSITATDD698 pKa = 3.47ASKK701 pKa = 11.53LEE703 pKa = 4.62GNTGTTPFTFTVSRR717 pKa = 11.84TGGTADD723 pKa = 5.52AITLAYY729 pKa = 10.16AVTGSGASAADD740 pKa = 3.37AADD743 pKa = 3.84FGGTLPSGTVTIAAGQASTTLMINVTGDD771 pKa = 3.65GLVEE775 pKa = 4.82ADD777 pKa = 4.89DD778 pKa = 4.8GFTVTLSNATANATIVQSTAIGTIRR803 pKa = 11.84NDD805 pKa = 3.56DD806 pKa = 3.85QPPYY810 pKa = 9.62TPPTAPTAVADD821 pKa = 4.06TGSVTLGTALTKK833 pKa = 10.4AAPGVLANDD842 pKa = 3.88SGAGATLTVTAVNGQAANVGKK863 pKa = 10.15ALAGAYY869 pKa = 10.02GNLTLNADD877 pKa = 3.71GSYY880 pKa = 10.76SYY882 pKa = 11.67APDD885 pKa = 3.24FAKK888 pKa = 10.6AVFTGSVVDD897 pKa = 3.81HH898 pKa = 6.31FTYY901 pKa = 10.8TDD903 pKa = 3.37TANGQTATTSLDD915 pKa = 3.14IAVAPPSAATLALFGTVVTNPASQTGGIYY944 pKa = 10.73GLYY947 pKa = 8.79TALLNRR953 pKa = 11.84VPDD956 pKa = 3.98ALGLEE961 pKa = 4.58GFSAAIQAGSDD972 pKa = 3.47LTTVASALLGSAEE985 pKa = 4.15RR986 pKa = 11.84GGTVSDD992 pKa = 3.42PTTYY996 pKa = 10.84VQGLYY1001 pKa = 11.02ANALHH1006 pKa = 6.13RR1007 pKa = 11.84TADD1010 pKa = 3.37AGGLNFFVNEE1020 pKa = 3.95LNAGVSQATVAVQIATSSEE1039 pKa = 3.98AQSLNTPAFQRR1050 pKa = 11.84GVFVTDD1056 pKa = 5.03AIDD1059 pKa = 3.51AGVARR1064 pKa = 11.84LYY1066 pKa = 10.98YY1067 pKa = 10.66GLLNRR1072 pKa = 11.84SPDD1075 pKa = 3.44AGGLGSFEE1083 pKa = 4.43GLVKK1087 pKa = 10.23QAAASGAAAGAIQGLATVANVMLGSPEE1114 pKa = 3.77FAATHH1119 pKa = 6.72AGQTSAAYY1127 pKa = 10.27VDD1129 pKa = 3.93SLYY1132 pKa = 11.25VGALGRR1138 pKa = 11.84HH1139 pKa = 6.1ADD1141 pKa = 3.52AAGASYY1147 pKa = 10.32FGAEE1151 pKa = 4.13LAQGVSRR1158 pKa = 11.84ATVALQIVEE1167 pKa = 4.19SAEE1170 pKa = 4.13AQVHH1174 pKa = 4.62LVGVIEE1180 pKa = 5.23QGFQLTAA1187 pKa = 3.68
MM1 pKa = 7.79PATFGTATTTLVEE14 pKa = 4.24SEE16 pKa = 4.03PWEE19 pKa = 4.45LSAADD24 pKa = 4.13LNGDD28 pKa = 3.67GKK30 pKa = 11.33LDD32 pKa = 3.47LVVPNTASNSLSVLIGNGDD51 pKa = 3.52GSFRR55 pKa = 11.84PAVNYY60 pKa = 8.21PVSSSPISGALADD73 pKa = 3.65VTGDD77 pKa = 3.47GKK79 pKa = 10.64IDD81 pKa = 3.29IVVGINGAPSLLAGNGDD98 pKa = 3.84GTFQAAAGIAGGNALQSYY116 pKa = 7.12WLRR119 pKa = 11.84AVDD122 pKa = 4.87LNADD126 pKa = 3.79GKK128 pKa = 11.42LDD130 pKa = 3.79LAYY133 pKa = 9.96TNFGPNQLVVQYY145 pKa = 10.94GNGDD149 pKa = 3.6GTFGTGALYY158 pKa = 10.66SVGNLPRR165 pKa = 11.84APAIGDD171 pKa = 3.65FNHH174 pKa = 7.23DD175 pKa = 3.52GKK177 pKa = 11.43LDD179 pKa = 3.41IVTANSGATSVSVLLANADD198 pKa = 3.33GTYY201 pKa = 10.24QSQVPYY207 pKa = 9.99PVGASPYY214 pKa = 10.39SVATGDD220 pKa = 3.89FNGDD224 pKa = 3.14GNLDD228 pKa = 3.77LVTAALNANSYY239 pKa = 10.94SVLLGRR245 pKa = 11.84GDD247 pKa = 3.8GTFQPALTTAAQGNSNEE264 pKa = 4.06ITVADD269 pKa = 3.54VDD271 pKa = 4.22GDD273 pKa = 4.03GKK275 pKa = 11.2LDD277 pKa = 4.58LIIANSSANQVGIAYY292 pKa = 8.89GQGDD296 pKa = 4.18GTFSAWQTLPTGGGPYY312 pKa = 9.1TAIAVDD318 pKa = 4.05LNGDD322 pKa = 3.41GRR324 pKa = 11.84LDD326 pKa = 3.54LAVTNRR332 pKa = 11.84VSGTVTTYY340 pKa = 8.83MQSTPPTLSITGATVVEE357 pKa = 4.96GNSGTADD364 pKa = 3.11AAFVVTRR371 pKa = 11.84SDD373 pKa = 3.48TTGSPTATYY382 pKa = 9.2TITNSTTDD390 pKa = 3.35GADD393 pKa = 3.5FGPGFQATGTVTFAAGATTATINVPIRR420 pKa = 11.84GDD422 pKa = 3.52TLYY425 pKa = 10.57EE426 pKa = 3.92ASEE429 pKa = 4.49TFTVALSNPQGATISGTGQATGTITNDD456 pKa = 3.36DD457 pKa = 4.11APPTVTLSGGTSIAEE472 pKa = 4.42GNSGTTVVPITATLSAVSGLATTVTLAFGGTATQGTDD509 pKa = 3.32YY510 pKa = 9.81TVSSTTITIPAGSLSGSVNVNVLGDD535 pKa = 3.65TAVEE539 pKa = 4.16PNGTATVTIATATNATPAGAAQTVTIQNDD568 pKa = 3.65DD569 pKa = 3.0VSYY572 pKa = 11.17AITASAAAVAEE583 pKa = 4.65GSSGAGNAVTYY594 pKa = 9.13TVTRR598 pKa = 11.84TGDD601 pKa = 3.17MTQAGVATVRR611 pKa = 11.84LSGEE615 pKa = 3.81ATRR618 pKa = 11.84GDD620 pKa = 4.46DD621 pKa = 4.38YY622 pKa = 11.55STTLPANGQINFVAGDD638 pKa = 3.53ASKK641 pKa = 9.48TFTVTTVPDD650 pKa = 3.57TTVEE654 pKa = 3.91PNEE657 pKa = 4.07RR658 pKa = 11.84VIATITGVSGQGGATIGTPASATTTITNDD687 pKa = 3.13DD688 pKa = 3.75QYY690 pKa = 11.49GVSITATDD698 pKa = 3.47ASKK701 pKa = 11.53LEE703 pKa = 4.62GNTGTTPFTFTVSRR717 pKa = 11.84TGGTADD723 pKa = 5.52AITLAYY729 pKa = 10.16AVTGSGASAADD740 pKa = 3.37AADD743 pKa = 3.84FGGTLPSGTVTIAAGQASTTLMINVTGDD771 pKa = 3.65GLVEE775 pKa = 4.82ADD777 pKa = 4.89DD778 pKa = 4.8GFTVTLSNATANATIVQSTAIGTIRR803 pKa = 11.84NDD805 pKa = 3.56DD806 pKa = 3.85QPPYY810 pKa = 9.62TPPTAPTAVADD821 pKa = 4.06TGSVTLGTALTKK833 pKa = 10.4AAPGVLANDD842 pKa = 3.88SGAGATLTVTAVNGQAANVGKK863 pKa = 10.15ALAGAYY869 pKa = 10.02GNLTLNADD877 pKa = 3.71GSYY880 pKa = 10.76SYY882 pKa = 11.67APDD885 pKa = 3.24FAKK888 pKa = 10.6AVFTGSVVDD897 pKa = 3.81HH898 pKa = 6.31FTYY901 pKa = 10.8TDD903 pKa = 3.37TANGQTATTSLDD915 pKa = 3.14IAVAPPSAATLALFGTVVTNPASQTGGIYY944 pKa = 10.73GLYY947 pKa = 8.79TALLNRR953 pKa = 11.84VPDD956 pKa = 3.98ALGLEE961 pKa = 4.58GFSAAIQAGSDD972 pKa = 3.47LTTVASALLGSAEE985 pKa = 4.15RR986 pKa = 11.84GGTVSDD992 pKa = 3.42PTTYY996 pKa = 10.84VQGLYY1001 pKa = 11.02ANALHH1006 pKa = 6.13RR1007 pKa = 11.84TADD1010 pKa = 3.37AGGLNFFVNEE1020 pKa = 3.95LNAGVSQATVAVQIATSSEE1039 pKa = 3.98AQSLNTPAFQRR1050 pKa = 11.84GVFVTDD1056 pKa = 5.03AIDD1059 pKa = 3.51AGVARR1064 pKa = 11.84LYY1066 pKa = 10.98YY1067 pKa = 10.66GLLNRR1072 pKa = 11.84SPDD1075 pKa = 3.44AGGLGSFEE1083 pKa = 4.43GLVKK1087 pKa = 10.23QAAASGAAAGAIQGLATVANVMLGSPEE1114 pKa = 3.77FAATHH1119 pKa = 6.72AGQTSAAYY1127 pKa = 10.27VDD1129 pKa = 3.93SLYY1132 pKa = 11.25VGALGRR1138 pKa = 11.84HH1139 pKa = 6.1ADD1141 pKa = 3.52AAGASYY1147 pKa = 10.32FGAEE1151 pKa = 4.13LAQGVSRR1158 pKa = 11.84ATVALQIVEE1167 pKa = 4.19SAEE1170 pKa = 4.13AQVHH1174 pKa = 4.62LVGVIEE1180 pKa = 5.23QGFQLTAA1187 pKa = 3.68
Molecular weight: 117.29 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2U8VM32|A0A2U8VM32_9RHIZ Uncharacterized protein OS=Methylobacterium sp. 17Sr1-43 OX=2202828 GN=DK427_01670 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATAGGRR28 pKa = 11.84KK29 pKa = 9.12VIARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AHH37 pKa = 4.95GRR39 pKa = 11.84KK40 pKa = 9.25RR41 pKa = 11.84LSAA44 pKa = 4.01
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATAGGRR28 pKa = 11.84KK29 pKa = 9.12VIARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AHH37 pKa = 4.95GRR39 pKa = 11.84KK40 pKa = 9.25RR41 pKa = 11.84LSAA44 pKa = 4.01
Molecular weight: 5.17 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1526070 |
29 |
2669 |
310.2 |
33.28 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.602 ± 0.062 | 0.844 ± 0.01 |
5.458 ± 0.024 | 5.691 ± 0.035 |
3.303 ± 0.023 | 9.18 ± 0.033 |
1.915 ± 0.017 | 4.206 ± 0.029 |
2.366 ± 0.031 | 10.542 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.994 ± 0.015 | 2.055 ± 0.019 |
5.909 ± 0.034 | 2.773 ± 0.019 |
8.483 ± 0.04 | 4.656 ± 0.021 |
5.164 ± 0.027 | 7.612 ± 0.028 |
1.233 ± 0.015 | 2.012 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
