
Pelagibaca abyssi
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacte
Average proteome isoelectric point is 6.16
Get precalculated fractions of proteins
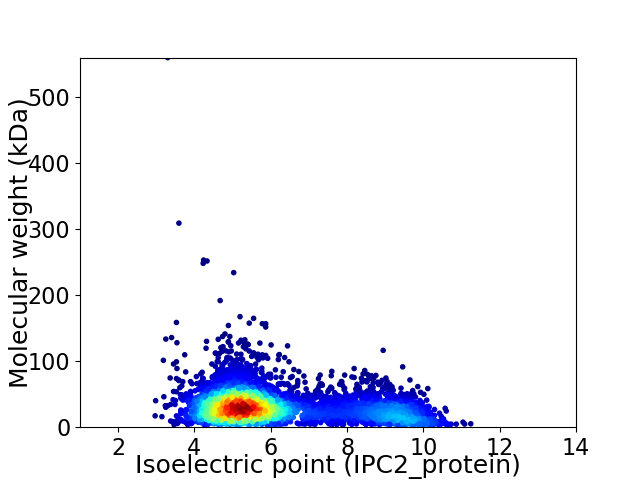
Virtual 2D-PAGE plot for 5048 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
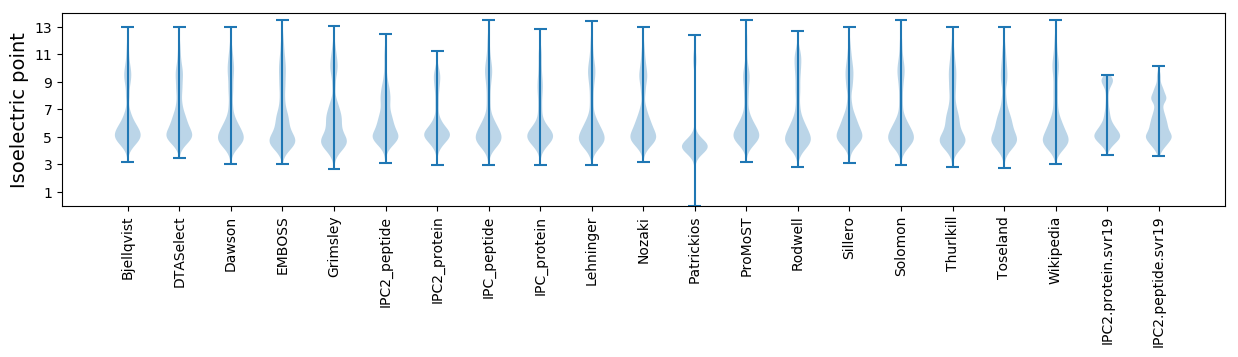
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1P8UWP2|A0A1P8UWP2_9RHOB Phage protein OS=Pelagibaca abyssi OX=1250539 GN=Ga0080574_TMP3422 PE=4 SV=1
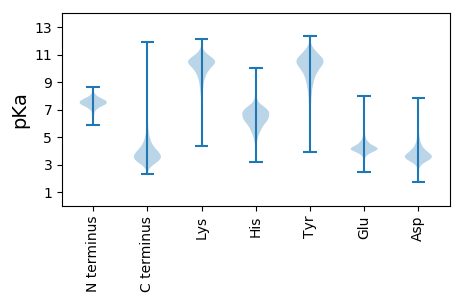
MM1 pKa = 7.82TITNDD6 pKa = 2.85NDD8 pKa = 3.65VIFGSGEE15 pKa = 4.35GYY17 pKa = 10.51FGRR20 pKa = 11.84SGTNPVSTVLSDD32 pKa = 3.6GRR34 pKa = 11.84IAVAWNEE41 pKa = 3.76YY42 pKa = 9.86QYY44 pKa = 10.94PGDD47 pKa = 3.83GSIPGIDD54 pKa = 3.64FDD56 pKa = 3.49TWTRR60 pKa = 11.84ILNADD65 pKa = 3.7GTPATEE71 pKa = 4.04AMLVNDD77 pKa = 4.07TRR79 pKa = 11.84VGSHH83 pKa = 6.48ASAEE87 pKa = 4.08ITALSDD93 pKa = 2.89GGYY96 pKa = 9.25VIGWITMNRR105 pKa = 11.84ATVEE109 pKa = 4.12GGTQTVYY116 pKa = 10.44TYY118 pKa = 10.21DD119 pKa = 2.91AHH121 pKa = 6.64VRR123 pKa = 11.84SFTAAGAPEE132 pKa = 4.75GPAVLVSPDD141 pKa = 3.52LEE143 pKa = 4.65TYY145 pKa = 10.85DD146 pKa = 3.75PADD149 pKa = 3.68YY150 pKa = 11.39NSILASNIEE159 pKa = 3.95NVNIVSLAAGGAVVIYY175 pKa = 8.43EE176 pKa = 3.98YY177 pKa = 10.79RR178 pKa = 11.84GGGSGYY184 pKa = 10.62SYY186 pKa = 11.25GGTHH190 pKa = 6.61ARR192 pKa = 11.84IVGNDD197 pKa = 3.71GQSLGEE203 pKa = 3.96PVRR206 pKa = 11.84VWEE209 pKa = 4.81GGLFNLDD216 pKa = 3.15AVQLSNGDD224 pKa = 3.62LAFVEE229 pKa = 4.95FAGDD233 pKa = 3.59LSGYY237 pKa = 8.41RR238 pKa = 11.84VRR240 pKa = 11.84LSAADD245 pKa = 3.89LTSAPATIAGASGPVVIDD263 pKa = 4.56HH264 pKa = 7.6DD265 pKa = 4.19PQPDD269 pKa = 3.56QNAGGYY275 pKa = 10.07GSAPRR280 pKa = 11.84IAALSDD286 pKa = 2.96GGFAVLYY293 pKa = 9.77GYY295 pKa = 10.22NVQLGADD302 pKa = 4.01DD303 pKa = 5.44DD304 pKa = 4.32EE305 pKa = 5.04SLRR308 pKa = 11.84IDD310 pKa = 4.06RR311 pKa = 11.84FDD313 pKa = 3.5AQGGYY318 pKa = 10.0QSTVSIPVPDD328 pKa = 4.49DD329 pKa = 3.29TVQQPGAIPYY339 pKa = 8.78EE340 pKa = 3.98ILEE343 pKa = 4.15LSGNRR348 pKa = 11.84LLVAWSHH355 pKa = 4.46VVAYY359 pKa = 10.41GDD361 pKa = 3.66TDD363 pKa = 3.08IMAVVINADD372 pKa = 3.49GSLDD376 pKa = 3.63SAPAVINPNTANYY389 pKa = 9.75QLLGDD394 pKa = 4.57LTALPDD400 pKa = 3.68GDD402 pKa = 4.39VFMSLADD409 pKa = 3.56TSGVQVGGVADD420 pKa = 3.82YY421 pKa = 9.99MHH423 pKa = 7.22GLFLGMPEE431 pKa = 4.54DD432 pKa = 4.41PGANTPVVRR441 pKa = 11.84IGTAGDD447 pKa = 3.51DD448 pKa = 3.76RR449 pKa = 11.84LVGTANNDD457 pKa = 3.56TLRR460 pKa = 11.84GLDD463 pKa = 3.91GNDD466 pKa = 3.24TLEE469 pKa = 4.62GRR471 pKa = 11.84DD472 pKa = 4.12GDD474 pKa = 4.04DD475 pKa = 3.35QLFGGDD481 pKa = 4.93DD482 pKa = 3.66NDD484 pKa = 4.54LVLGGDD490 pKa = 4.45GSDD493 pKa = 3.22TMAGDD498 pKa = 4.2GGNDD502 pKa = 3.45TLRR505 pKa = 11.84GGNSAADD512 pKa = 4.03LRR514 pKa = 11.84DD515 pKa = 3.69VIYY518 pKa = 10.74GGAGDD523 pKa = 4.9DD524 pKa = 5.51DD525 pKa = 4.46IDD527 pKa = 4.39GGYY530 pKa = 11.11GNDD533 pKa = 4.01EE534 pKa = 4.14LRR536 pKa = 11.84GDD538 pKa = 4.22AGNDD542 pKa = 3.85TIVGGYY548 pKa = 9.71GADD551 pKa = 3.37TVIGGTGNDD560 pKa = 3.66ALTGQTWSDD569 pKa = 3.87AIFGGDD575 pKa = 3.55GMDD578 pKa = 5.06FINGGFGHH586 pKa = 7.05DD587 pKa = 3.55RR588 pKa = 11.84VNGGTGADD596 pKa = 3.08RR597 pKa = 11.84FYY599 pKa = 11.0HH600 pKa = 6.38LGVEE604 pKa = 4.6GHH606 pKa = 6.22GSDD609 pKa = 4.17WIQDD613 pKa = 3.69FSDD616 pKa = 3.79EE617 pKa = 4.33EE618 pKa = 5.54GDD620 pKa = 3.79LLVFGGTGAAVEE632 pKa = 4.15DD633 pKa = 4.24FQVNFTEE640 pKa = 4.42TANAGAAGVEE650 pKa = 4.33EE651 pKa = 4.43TFVIYY656 pKa = 10.48RR657 pKa = 11.84PTGQILWALVDD668 pKa = 4.72GGAQDD673 pKa = 5.28EE674 pKa = 5.07IMLQLDD680 pKa = 3.58GMSYY684 pKa = 11.5DD685 pKa = 4.04LLAA688 pKa = 5.6
MM1 pKa = 7.82TITNDD6 pKa = 2.85NDD8 pKa = 3.65VIFGSGEE15 pKa = 4.35GYY17 pKa = 10.51FGRR20 pKa = 11.84SGTNPVSTVLSDD32 pKa = 3.6GRR34 pKa = 11.84IAVAWNEE41 pKa = 3.76YY42 pKa = 9.86QYY44 pKa = 10.94PGDD47 pKa = 3.83GSIPGIDD54 pKa = 3.64FDD56 pKa = 3.49TWTRR60 pKa = 11.84ILNADD65 pKa = 3.7GTPATEE71 pKa = 4.04AMLVNDD77 pKa = 4.07TRR79 pKa = 11.84VGSHH83 pKa = 6.48ASAEE87 pKa = 4.08ITALSDD93 pKa = 2.89GGYY96 pKa = 9.25VIGWITMNRR105 pKa = 11.84ATVEE109 pKa = 4.12GGTQTVYY116 pKa = 10.44TYY118 pKa = 10.21DD119 pKa = 2.91AHH121 pKa = 6.64VRR123 pKa = 11.84SFTAAGAPEE132 pKa = 4.75GPAVLVSPDD141 pKa = 3.52LEE143 pKa = 4.65TYY145 pKa = 10.85DD146 pKa = 3.75PADD149 pKa = 3.68YY150 pKa = 11.39NSILASNIEE159 pKa = 3.95NVNIVSLAAGGAVVIYY175 pKa = 8.43EE176 pKa = 3.98YY177 pKa = 10.79RR178 pKa = 11.84GGGSGYY184 pKa = 10.62SYY186 pKa = 11.25GGTHH190 pKa = 6.61ARR192 pKa = 11.84IVGNDD197 pKa = 3.71GQSLGEE203 pKa = 3.96PVRR206 pKa = 11.84VWEE209 pKa = 4.81GGLFNLDD216 pKa = 3.15AVQLSNGDD224 pKa = 3.62LAFVEE229 pKa = 4.95FAGDD233 pKa = 3.59LSGYY237 pKa = 8.41RR238 pKa = 11.84VRR240 pKa = 11.84LSAADD245 pKa = 3.89LTSAPATIAGASGPVVIDD263 pKa = 4.56HH264 pKa = 7.6DD265 pKa = 4.19PQPDD269 pKa = 3.56QNAGGYY275 pKa = 10.07GSAPRR280 pKa = 11.84IAALSDD286 pKa = 2.96GGFAVLYY293 pKa = 9.77GYY295 pKa = 10.22NVQLGADD302 pKa = 4.01DD303 pKa = 5.44DD304 pKa = 4.32EE305 pKa = 5.04SLRR308 pKa = 11.84IDD310 pKa = 4.06RR311 pKa = 11.84FDD313 pKa = 3.5AQGGYY318 pKa = 10.0QSTVSIPVPDD328 pKa = 4.49DD329 pKa = 3.29TVQQPGAIPYY339 pKa = 8.78EE340 pKa = 3.98ILEE343 pKa = 4.15LSGNRR348 pKa = 11.84LLVAWSHH355 pKa = 4.46VVAYY359 pKa = 10.41GDD361 pKa = 3.66TDD363 pKa = 3.08IMAVVINADD372 pKa = 3.49GSLDD376 pKa = 3.63SAPAVINPNTANYY389 pKa = 9.75QLLGDD394 pKa = 4.57LTALPDD400 pKa = 3.68GDD402 pKa = 4.39VFMSLADD409 pKa = 3.56TSGVQVGGVADD420 pKa = 3.82YY421 pKa = 9.99MHH423 pKa = 7.22GLFLGMPEE431 pKa = 4.54DD432 pKa = 4.41PGANTPVVRR441 pKa = 11.84IGTAGDD447 pKa = 3.51DD448 pKa = 3.76RR449 pKa = 11.84LVGTANNDD457 pKa = 3.56TLRR460 pKa = 11.84GLDD463 pKa = 3.91GNDD466 pKa = 3.24TLEE469 pKa = 4.62GRR471 pKa = 11.84DD472 pKa = 4.12GDD474 pKa = 4.04DD475 pKa = 3.35QLFGGDD481 pKa = 4.93DD482 pKa = 3.66NDD484 pKa = 4.54LVLGGDD490 pKa = 4.45GSDD493 pKa = 3.22TMAGDD498 pKa = 4.2GGNDD502 pKa = 3.45TLRR505 pKa = 11.84GGNSAADD512 pKa = 4.03LRR514 pKa = 11.84DD515 pKa = 3.69VIYY518 pKa = 10.74GGAGDD523 pKa = 4.9DD524 pKa = 5.51DD525 pKa = 4.46IDD527 pKa = 4.39GGYY530 pKa = 11.11GNDD533 pKa = 4.01EE534 pKa = 4.14LRR536 pKa = 11.84GDD538 pKa = 4.22AGNDD542 pKa = 3.85TIVGGYY548 pKa = 9.71GADD551 pKa = 3.37TVIGGTGNDD560 pKa = 3.66ALTGQTWSDD569 pKa = 3.87AIFGGDD575 pKa = 3.55GMDD578 pKa = 5.06FINGGFGHH586 pKa = 7.05DD587 pKa = 3.55RR588 pKa = 11.84VNGGTGADD596 pKa = 3.08RR597 pKa = 11.84FYY599 pKa = 11.0HH600 pKa = 6.38LGVEE604 pKa = 4.6GHH606 pKa = 6.22GSDD609 pKa = 4.17WIQDD613 pKa = 3.69FSDD616 pKa = 3.79EE617 pKa = 4.33EE618 pKa = 5.54GDD620 pKa = 3.79LLVFGGTGAAVEE632 pKa = 4.15DD633 pKa = 4.24FQVNFTEE640 pKa = 4.42TANAGAAGVEE650 pKa = 4.33EE651 pKa = 4.43TFVIYY656 pKa = 10.48RR657 pKa = 11.84PTGQILWALVDD668 pKa = 4.72GGAQDD673 pKa = 5.28EE674 pKa = 5.07IMLQLDD680 pKa = 3.58GMSYY684 pKa = 11.5DD685 pKa = 4.04LLAA688 pKa = 5.6
Molecular weight: 71.14 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1P8UVF7|A0A1P8UVF7_9RHOB Uncharacterized protein OS=Pelagibaca abyssi OX=1250539 GN=Ga0080574_TMP3028 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.44AGRR28 pKa = 11.84KK29 pKa = 8.46VLNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.51QLSAA44 pKa = 3.97
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.44AGRR28 pKa = 11.84KK29 pKa = 8.46VLNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.51QLSAA44 pKa = 3.97
Molecular weight: 5.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1524221 |
37 |
5509 |
301.9 |
32.7 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.814 ± 0.053 | 0.878 ± 0.012 |
5.932 ± 0.041 | 6.359 ± 0.039 |
3.577 ± 0.019 | 8.949 ± 0.045 |
2.009 ± 0.018 | 4.864 ± 0.025 |
2.808 ± 0.032 | 10.249 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.723 ± 0.021 | 2.322 ± 0.021 |
5.258 ± 0.029 | 3.046 ± 0.017 |
7.092 ± 0.041 | 5.1 ± 0.028 |
5.308 ± 0.033 | 7.119 ± 0.028 |
1.39 ± 0.014 | 2.202 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
