
Vibrio phage CKB-S1
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Myoviridae; unclassified Myoviridae
Average proteome isoelectric point is 6.08
Get precalculated fractions of proteins
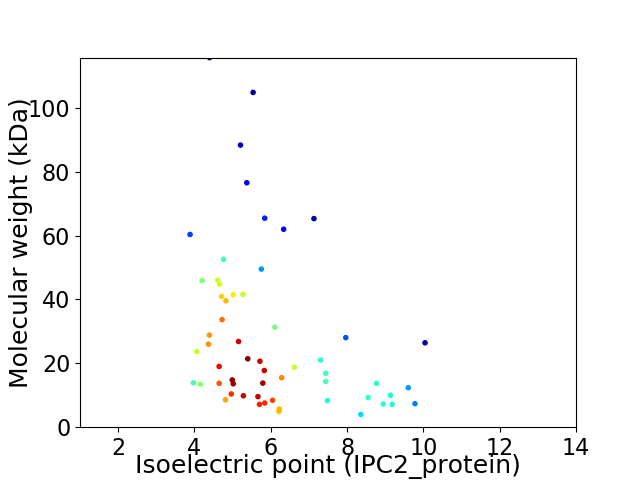
Virtual 2D-PAGE plot for 58 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
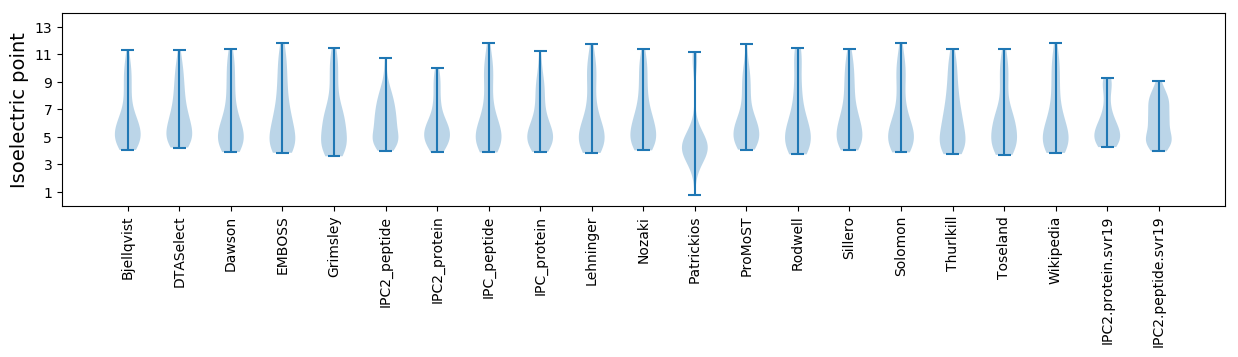
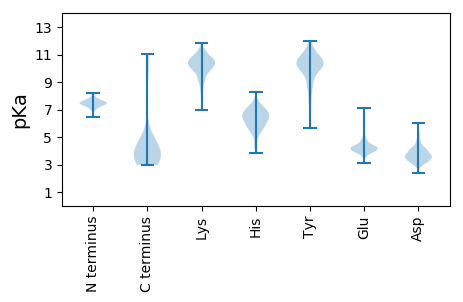
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1E1GE49|A0A1E1GE49_9CAUD Uncharacterized protein OS=Vibrio phage CKB-S1 OX=1903259 PE=4 SV=1
MM1 pKa = 7.73LIKK4 pKa = 10.0LTRR7 pKa = 11.84ATRR10 pKa = 11.84YY11 pKa = 9.82LGAVLNIGGAGGAGFVYY28 pKa = 10.06PDD30 pKa = 3.19SADD33 pKa = 3.64RR34 pKa = 11.84PFATIAARR42 pKa = 11.84NTWAAANLGDD52 pKa = 4.61LVDD55 pKa = 4.38GSTVVQVAGTPATWYY70 pKa = 9.68LWTGEE75 pKa = 4.29TSPSSHH81 pKa = 7.66DD82 pKa = 3.39SALWIDD88 pKa = 4.17ATPLIQGPAGEE99 pKa = 4.92DD100 pKa = 4.11GIDD103 pKa = 3.48GTALEE108 pKa = 4.63FTSLSARR115 pKa = 11.84DD116 pKa = 3.49SFFISRR122 pKa = 11.84PDD124 pKa = 3.34LLLNGLPVLVNIGNNTVGIYY144 pKa = 8.68TWTGPTNPSDD154 pKa = 4.22LVASDD159 pKa = 4.04TRR161 pKa = 11.84WSQASQAVPNNSLFVGSTRR180 pKa = 11.84VSNAVEE186 pKa = 4.15NIVAEE191 pKa = 4.37TADD194 pKa = 3.39GRR196 pKa = 11.84QFLATGVRR204 pKa = 11.84FDD206 pKa = 4.38DD207 pKa = 3.76TGSRR211 pKa = 11.84PFGFTEE217 pKa = 4.27LAAATTLNVNTANTQDD233 pKa = 3.58MPDD236 pKa = 4.11PFTVQYY242 pKa = 7.66TTFGDD247 pKa = 3.63NLTTDD252 pKa = 4.27FDD254 pKa = 5.28FIPAEE259 pKa = 4.15AGEE262 pKa = 3.99LRR264 pKa = 11.84AEE266 pKa = 4.37FFLGADD272 pKa = 3.85DD273 pKa = 3.9QAPLIFDD280 pKa = 3.32EE281 pKa = 4.9RR282 pKa = 11.84RR283 pKa = 11.84AVTQAEE289 pKa = 3.99VDD291 pKa = 3.15AGEE294 pKa = 4.24FVSFGVGNPYY304 pKa = 10.82LLDD307 pKa = 3.93EE308 pKa = 4.66GLQLFARR315 pKa = 11.84FSGVQLRR322 pKa = 11.84GGVVDD327 pKa = 4.9NPGTPLDD334 pKa = 3.95GQTIISFRR342 pKa = 11.84STVQAFTARR351 pKa = 11.84QVMRR355 pKa = 11.84ADD357 pKa = 3.82EE358 pKa = 4.27FTGAAVRR365 pKa = 11.84DD366 pKa = 4.21LLTALTGGDD375 pKa = 4.13RR376 pKa = 11.84LPASAIRR383 pKa = 11.84DD384 pKa = 3.66LPAGVTLRR392 pKa = 11.84TASEE396 pKa = 4.04TADD399 pKa = 3.82LLEE402 pKa = 4.67TLTGDD407 pKa = 4.17DD408 pKa = 4.13RR409 pKa = 11.84LAASAIRR416 pKa = 11.84DD417 pKa = 3.67LPADD421 pKa = 3.11VDD423 pKa = 3.91NFVDD427 pKa = 3.57QLTVGLSGQDD437 pKa = 3.22LTITLGRR444 pKa = 11.84TGTLPDD450 pKa = 4.05LSQTVTLPSGGGTDD464 pKa = 4.17PGPGPDD470 pKa = 2.96VLYY473 pKa = 10.72YY474 pKa = 10.84GRR476 pKa = 11.84STSNNPATVDD486 pKa = 3.98LATLTQIDD494 pKa = 4.07PTDD497 pKa = 4.05PQTVSTGVAVAGDD510 pKa = 3.56YY511 pKa = 10.75FIILTANTHH520 pKa = 7.17DD521 pKa = 3.43ITTITDD527 pKa = 3.33TVLQQDD533 pKa = 3.92VTSLFTKK540 pKa = 10.28TEE542 pKa = 3.96DD543 pKa = 3.34VRR545 pKa = 11.84AEE547 pKa = 4.06GAVTLDD553 pKa = 3.76SYY555 pKa = 11.96VIGPLNAGVNEE566 pKa = 4.09QYY568 pKa = 11.28VLEE571 pKa = 4.45FF572 pKa = 4.38
MM1 pKa = 7.73LIKK4 pKa = 10.0LTRR7 pKa = 11.84ATRR10 pKa = 11.84YY11 pKa = 9.82LGAVLNIGGAGGAGFVYY28 pKa = 10.06PDD30 pKa = 3.19SADD33 pKa = 3.64RR34 pKa = 11.84PFATIAARR42 pKa = 11.84NTWAAANLGDD52 pKa = 4.61LVDD55 pKa = 4.38GSTVVQVAGTPATWYY70 pKa = 9.68LWTGEE75 pKa = 4.29TSPSSHH81 pKa = 7.66DD82 pKa = 3.39SALWIDD88 pKa = 4.17ATPLIQGPAGEE99 pKa = 4.92DD100 pKa = 4.11GIDD103 pKa = 3.48GTALEE108 pKa = 4.63FTSLSARR115 pKa = 11.84DD116 pKa = 3.49SFFISRR122 pKa = 11.84PDD124 pKa = 3.34LLLNGLPVLVNIGNNTVGIYY144 pKa = 8.68TWTGPTNPSDD154 pKa = 4.22LVASDD159 pKa = 4.04TRR161 pKa = 11.84WSQASQAVPNNSLFVGSTRR180 pKa = 11.84VSNAVEE186 pKa = 4.15NIVAEE191 pKa = 4.37TADD194 pKa = 3.39GRR196 pKa = 11.84QFLATGVRR204 pKa = 11.84FDD206 pKa = 4.38DD207 pKa = 3.76TGSRR211 pKa = 11.84PFGFTEE217 pKa = 4.27LAAATTLNVNTANTQDD233 pKa = 3.58MPDD236 pKa = 4.11PFTVQYY242 pKa = 7.66TTFGDD247 pKa = 3.63NLTTDD252 pKa = 4.27FDD254 pKa = 5.28FIPAEE259 pKa = 4.15AGEE262 pKa = 3.99LRR264 pKa = 11.84AEE266 pKa = 4.37FFLGADD272 pKa = 3.85DD273 pKa = 3.9QAPLIFDD280 pKa = 3.32EE281 pKa = 4.9RR282 pKa = 11.84RR283 pKa = 11.84AVTQAEE289 pKa = 3.99VDD291 pKa = 3.15AGEE294 pKa = 4.24FVSFGVGNPYY304 pKa = 10.82LLDD307 pKa = 3.93EE308 pKa = 4.66GLQLFARR315 pKa = 11.84FSGVQLRR322 pKa = 11.84GGVVDD327 pKa = 4.9NPGTPLDD334 pKa = 3.95GQTIISFRR342 pKa = 11.84STVQAFTARR351 pKa = 11.84QVMRR355 pKa = 11.84ADD357 pKa = 3.82EE358 pKa = 4.27FTGAAVRR365 pKa = 11.84DD366 pKa = 4.21LLTALTGGDD375 pKa = 4.13RR376 pKa = 11.84LPASAIRR383 pKa = 11.84DD384 pKa = 3.66LPAGVTLRR392 pKa = 11.84TASEE396 pKa = 4.04TADD399 pKa = 3.82LLEE402 pKa = 4.67TLTGDD407 pKa = 4.17DD408 pKa = 4.13RR409 pKa = 11.84LAASAIRR416 pKa = 11.84DD417 pKa = 3.67LPADD421 pKa = 3.11VDD423 pKa = 3.91NFVDD427 pKa = 3.57QLTVGLSGQDD437 pKa = 3.22LTITLGRR444 pKa = 11.84TGTLPDD450 pKa = 4.05LSQTVTLPSGGGTDD464 pKa = 4.17PGPGPDD470 pKa = 2.96VLYY473 pKa = 10.72YY474 pKa = 10.84GRR476 pKa = 11.84STSNNPATVDD486 pKa = 3.98LATLTQIDD494 pKa = 4.07PTDD497 pKa = 4.05PQTVSTGVAVAGDD510 pKa = 3.56YY511 pKa = 10.75FIILTANTHH520 pKa = 7.17DD521 pKa = 3.43ITTITDD527 pKa = 3.33TVLQQDD533 pKa = 3.92VTSLFTKK540 pKa = 10.28TEE542 pKa = 3.96DD543 pKa = 3.34VRR545 pKa = 11.84AEE547 pKa = 4.06GAVTLDD553 pKa = 3.76SYY555 pKa = 11.96VIGPLNAGVNEE566 pKa = 4.09QYY568 pKa = 11.28VLEE571 pKa = 4.45FF572 pKa = 4.38
Molecular weight: 60.32 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1E1GEL5|A0A1E1GEL5_9CAUD Gp62 OS=Vibrio phage CKB-S1 OX=1903259 GN=BcepB1A gene62 PE=4 SV=1
MM1 pKa = 7.64ASCPHH6 pKa = 4.22THH8 pKa = 6.29FRR10 pKa = 11.84RR11 pKa = 11.84GTTVLVILKK20 pKa = 10.23DD21 pKa = 3.37STKK24 pKa = 10.15RR25 pKa = 11.84VCKK28 pKa = 10.34FKK30 pKa = 10.69EE31 pKa = 4.12SRR33 pKa = 11.84SKK35 pKa = 10.9HH36 pKa = 4.5IVLWDD41 pKa = 3.7GEE43 pKa = 4.43KK44 pKa = 10.69LPHH47 pKa = 6.45KK48 pKa = 10.25LIRR51 pKa = 11.84QVLIYY56 pKa = 9.91RR57 pKa = 11.84RR58 pKa = 11.84KK59 pKa = 10.05GGQAA63 pKa = 2.98
MM1 pKa = 7.64ASCPHH6 pKa = 4.22THH8 pKa = 6.29FRR10 pKa = 11.84RR11 pKa = 11.84GTTVLVILKK20 pKa = 10.23DD21 pKa = 3.37STKK24 pKa = 10.15RR25 pKa = 11.84VCKK28 pKa = 10.34FKK30 pKa = 10.69EE31 pKa = 4.12SRR33 pKa = 11.84SKK35 pKa = 10.9HH36 pKa = 4.5IVLWDD41 pKa = 3.7GEE43 pKa = 4.43KK44 pKa = 10.69LPHH47 pKa = 6.45KK48 pKa = 10.25LIRR51 pKa = 11.84QVLIYY56 pKa = 9.91RR57 pKa = 11.84RR58 pKa = 11.84KK59 pKa = 10.05GGQAA63 pKa = 2.98
Molecular weight: 7.36 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
15186 |
36 |
1064 |
261.8 |
28.71 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.641 ± 0.417 | 0.711 ± 0.154 |
6.249 ± 0.222 | 5.9 ± 0.269 |
3.464 ± 0.181 | 8.205 ± 0.188 |
1.791 ± 0.191 | 4.682 ± 0.22 |
3.674 ± 0.357 | 8.165 ± 0.239 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.496 ± 0.207 | 4.32 ± 0.213 |
4.748 ± 0.221 | 5.123 ± 0.511 |
6.091 ± 0.275 | 4.899 ± 0.174 |
6.934 ± 0.392 | 7.698 ± 0.258 |
1.468 ± 0.159 | 2.739 ± 0.159 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
