
BtMf-AlphaCoV/JX2012
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Nidovirales; Cornidovirineae; Coronaviridae; Orthocoronavirinae; Alphacoronavirus; Minunacovirus; Miniopterus bat coronavirus 1
Average proteome isoelectric point is 7.2
Get precalculated fractions of proteins
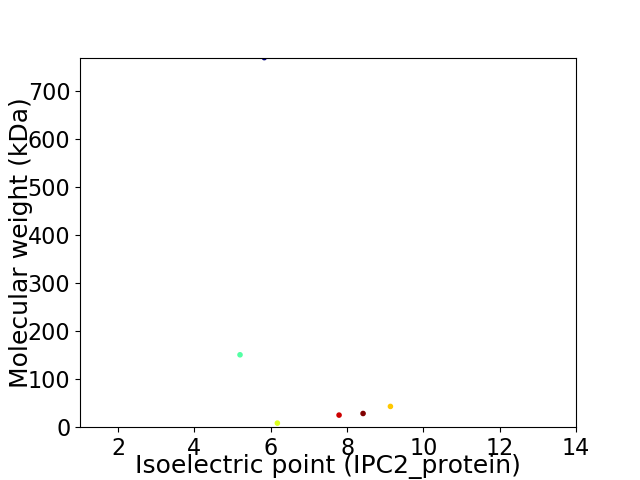
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
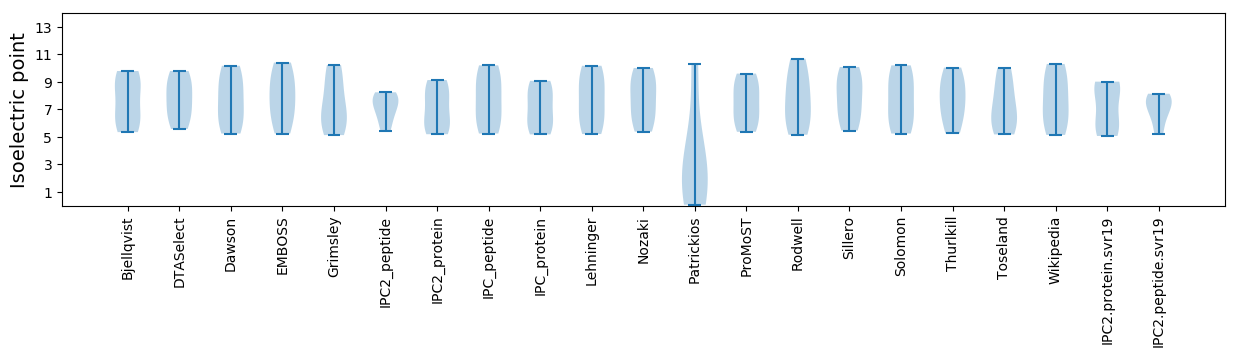
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0U1WHB9|A0A0U1WHB9_9ALPC Envelope small membrane protein OS=BtMf-AlphaCoV/JX2012 OX=1503288 GN=E PE=3 SV=1
MM1 pKa = 7.44FIIYY5 pKa = 9.43FCVALCINFASANIGCSSNGNLGMYY30 pKa = 9.8RR31 pKa = 11.84LKK33 pKa = 10.91LGLPNNVTNAYY44 pKa = 9.78VSGYY48 pKa = 10.59LPTPDD53 pKa = 4.14QWNCTQNSAGTANITYY69 pKa = 9.49NARR72 pKa = 11.84AVFISYY78 pKa = 10.44YY79 pKa = 10.66KK80 pKa = 10.44FGTGVAVGVGNSKK93 pKa = 11.17SNDD96 pKa = 2.93SFGIYY101 pKa = 10.01FYY103 pKa = 11.02HH104 pKa = 7.37HH105 pKa = 6.81NGEE108 pKa = 4.06QKK110 pKa = 9.88TILRR114 pKa = 11.84VCKK117 pKa = 9.73WNGRR121 pKa = 11.84YY122 pKa = 8.23MPSYY126 pKa = 10.52RR127 pKa = 11.84PTSTTNGYY135 pKa = 10.5DD136 pKa = 3.21CVINVNLNLRR146 pKa = 11.84FVDD149 pKa = 3.87QQSHH153 pKa = 5.85NDD155 pKa = 3.33VFGLSWSGDD164 pKa = 3.05RR165 pKa = 11.84LIIYY169 pKa = 10.05SVDD172 pKa = 3.21GVSSHH177 pKa = 6.6HH178 pKa = 6.76VDD180 pKa = 4.42GIGDD184 pKa = 4.25LDD186 pKa = 3.82VVSFRR191 pKa = 11.84CQLKK195 pKa = 7.94EE196 pKa = 4.08TCAHH200 pKa = 5.15QVVTKK205 pKa = 10.16PITAIVNTTSDD216 pKa = 3.69GFISGYY222 pKa = 7.31EE223 pKa = 3.86VCADD227 pKa = 3.28CDD229 pKa = 3.83GFPKK233 pKa = 10.72YY234 pKa = 10.73VFAVTDD240 pKa = 4.16GGAIPPNFQFDD251 pKa = 2.88NWFYY255 pKa = 10.18LTNSSTPISGMFTSVQPFNLSCVWPVPVLTSNSLPVHH292 pKa = 6.5FNMSLNNGAHH302 pKa = 6.45CNGYY306 pKa = 10.78SNLDD310 pKa = 3.5VGLVDD315 pKa = 5.3AMRR318 pKa = 11.84FSLNFTDD325 pKa = 3.65SQIIRR330 pKa = 11.84TGVISVITSTNVFNFSCTNSSVFGEE355 pKa = 4.38TNVIPFGDD363 pKa = 3.69VEE365 pKa = 4.23KK366 pKa = 9.67THH368 pKa = 6.5YY369 pKa = 10.86CFINYY374 pKa = 6.99FTNITTGEE382 pKa = 4.2SASQFVGILPPQVKK396 pKa = 9.71EE397 pKa = 3.69FVIMRR402 pKa = 11.84NGDD405 pKa = 3.34FHH407 pKa = 8.25LNGYY411 pKa = 10.09RR412 pKa = 11.84IFSVDD417 pKa = 3.14SVEE420 pKa = 4.69SVIFNISTKK429 pKa = 10.3DD430 pKa = 3.43GRR432 pKa = 11.84DD433 pKa = 3.2FWTVAFANNAEE444 pKa = 4.17VLTEE448 pKa = 4.01INATSIQNLLYY459 pKa = 10.82CNNPVNSIKK468 pKa = 10.55CQQLRR473 pKa = 11.84FNLDD477 pKa = 3.27DD478 pKa = 4.0GFYY481 pKa = 10.69SHH483 pKa = 7.19TFDD486 pKa = 5.35SVDD489 pKa = 3.28NLPRR493 pKa = 11.84TIVRR497 pKa = 11.84LPKK500 pKa = 10.35YY501 pKa = 8.13VTHH504 pKa = 6.86SIINVTVGINFEE516 pKa = 4.41NKK518 pKa = 8.8TLTNYY523 pKa = 10.83LIDD526 pKa = 4.39FAPDD530 pKa = 3.28DD531 pKa = 4.12VSIGKK536 pKa = 9.88DD537 pKa = 3.19NITTVCVDD545 pKa = 3.16TTTFTVRR552 pKa = 11.84LNVLAFSSFSTAVDD566 pKa = 3.57IQAGTCPFSYY576 pKa = 11.29NNLNNYY582 pKa = 9.99LGFGSLCFSTRR593 pKa = 11.84PNGGCMMSVVARR605 pKa = 11.84GYY607 pKa = 9.89FGEE610 pKa = 4.14FQKK613 pKa = 11.28VGVLYY618 pKa = 11.01VSFTKK623 pKa = 10.63GDD625 pKa = 3.57NVLGVPPNNVPTIGVSDD642 pKa = 3.7MSDD645 pKa = 2.83VKK647 pKa = 11.15LGVCTTFTIYY657 pKa = 10.65GHH659 pKa = 5.54TGRR662 pKa = 11.84GIINKK667 pKa = 9.73SNNTFVSGLFYY678 pKa = 10.84TSVTGNLLGFKK689 pKa = 10.45NSTTGEE695 pKa = 4.0IFSISPCQLTTQVAVISDD713 pKa = 4.37KK714 pKa = 11.0IVGVAAATTEE724 pKa = 4.1VKK726 pKa = 10.74LPFDD730 pKa = 4.24NYY732 pKa = 9.98VSLGSFYY739 pKa = 10.56YY740 pKa = 10.02HH741 pKa = 6.21YY742 pKa = 10.07RR743 pKa = 11.84NSSAEE748 pKa = 3.92LCKK751 pKa = 10.85NPSLMYY757 pKa = 10.57GGLGVCEE764 pKa = 5.02DD765 pKa = 3.57GRR767 pKa = 11.84LVNISRR773 pKa = 11.84SEE775 pKa = 4.02DD776 pKa = 3.45TFVPSAVISGNITIPANFSFVVQPEE801 pKa = 4.31YY802 pKa = 10.43IQIMTKK808 pKa = 9.87PVSVDD813 pKa = 2.99CSVYY817 pKa = 10.26VCNGNPRR824 pKa = 11.84CLQLLTQYY832 pKa = 11.57ASVCRR837 pKa = 11.84TVEE840 pKa = 4.29EE841 pKa = 4.13PLQLNARR848 pKa = 11.84LEE850 pKa = 4.16ALEE853 pKa = 4.37LTSMIIVQDD862 pKa = 3.48KK863 pKa = 8.43TLKK866 pKa = 10.86LGVASNFNDD875 pKa = 3.82TFDD878 pKa = 3.91LTLALPRR885 pKa = 11.84QHH887 pKa = 6.45QSRR890 pKa = 11.84SAIEE894 pKa = 4.54DD895 pKa = 3.65LLFSKK900 pKa = 10.43IVTSGLGTVDD910 pKa = 5.34DD911 pKa = 5.32DD912 pKa = 4.97YY913 pKa = 11.92KK914 pKa = 11.26EE915 pKa = 4.45CAAKK919 pKa = 9.16MANTIAEE926 pKa = 4.49AGCVQYY932 pKa = 11.5YY933 pKa = 9.78NGIMVLPGVVDD944 pKa = 4.33PSLLSQYY951 pKa = 9.05TAALTGAMVLGGVTAAAAIPFSLAVQARR979 pKa = 11.84LNYY982 pKa = 10.03LALQTDD988 pKa = 4.14VLQRR992 pKa = 11.84NQMILAQSFNAAMGNITVAFSGVSNAIQQTAQSLNTISQALNKK1035 pKa = 9.73VEE1037 pKa = 4.27NVVNEE1042 pKa = 3.91QGTALSQLTKK1052 pKa = 10.61QLASNFQAISSSIEE1066 pKa = 3.69DD1067 pKa = 3.97LYY1069 pKa = 11.84NRR1071 pKa = 11.84LDD1073 pKa = 3.6TLEE1076 pKa = 5.38ADD1078 pKa = 3.74QQVDD1082 pKa = 3.46RR1083 pKa = 11.84LITGRR1088 pKa = 11.84LAALNAFVTQQLLRR1102 pKa = 11.84YY1103 pKa = 8.9SEE1105 pKa = 4.28VRR1107 pKa = 11.84ASRR1110 pKa = 11.84QLAQEE1115 pKa = 4.75KK1116 pKa = 10.45INEE1119 pKa = 4.44CVKK1122 pKa = 10.53SQSSRR1127 pKa = 11.84YY1128 pKa = 8.35GFCGNGTHH1136 pKa = 5.87VFSVANAAPDD1146 pKa = 4.45GIMFLHH1152 pKa = 6.91ANLVPTAFIEE1162 pKa = 4.41VSAFAGVCVEE1172 pKa = 3.85GRR1174 pKa = 11.84ALVLRR1179 pKa = 11.84GRR1181 pKa = 11.84DD1182 pKa = 3.0EE1183 pKa = 4.97VLFQKK1188 pKa = 10.29PNTMQYY1194 pKa = 10.94LITPRR1199 pKa = 11.84LLFEE1203 pKa = 4.25PRR1205 pKa = 11.84VPVSADD1211 pKa = 3.25FVEE1214 pKa = 4.87VSSCNVTFINLTINEE1229 pKa = 4.53LPEE1232 pKa = 4.46LLPDD1236 pKa = 4.18YY1237 pKa = 10.98IDD1239 pKa = 3.45VNKK1242 pKa = 9.25TLKK1245 pKa = 10.34EE1246 pKa = 3.88FAATIPNRR1254 pKa = 11.84TEE1256 pKa = 3.59MQLTLNTYY1264 pKa = 9.78NATVLNLTDD1273 pKa = 3.74EE1274 pKa = 4.85VKK1276 pKa = 10.99SLMSQADD1283 pKa = 4.45DD1284 pKa = 3.54LTKK1287 pKa = 10.24IASEE1291 pKa = 4.28LNLTISKK1298 pKa = 10.27INNTLVEE1305 pKa = 4.51LEE1307 pKa = 3.89WLNRR1311 pKa = 11.84VEE1313 pKa = 5.97TYY1315 pKa = 10.4IKK1317 pKa = 9.3WPWYY1321 pKa = 7.96VWLAIAVTLIILVGLMLWCCLATGCCGCCSCLVNSCSDD1359 pKa = 3.33CGGRR1363 pKa = 11.84RR1364 pKa = 11.84LQRR1367 pKa = 11.84YY1368 pKa = 8.12EE1369 pKa = 3.64IEE1371 pKa = 4.3KK1372 pKa = 10.33VHH1374 pKa = 5.94IQQ1376 pKa = 3.19
MM1 pKa = 7.44FIIYY5 pKa = 9.43FCVALCINFASANIGCSSNGNLGMYY30 pKa = 9.8RR31 pKa = 11.84LKK33 pKa = 10.91LGLPNNVTNAYY44 pKa = 9.78VSGYY48 pKa = 10.59LPTPDD53 pKa = 4.14QWNCTQNSAGTANITYY69 pKa = 9.49NARR72 pKa = 11.84AVFISYY78 pKa = 10.44YY79 pKa = 10.66KK80 pKa = 10.44FGTGVAVGVGNSKK93 pKa = 11.17SNDD96 pKa = 2.93SFGIYY101 pKa = 10.01FYY103 pKa = 11.02HH104 pKa = 7.37HH105 pKa = 6.81NGEE108 pKa = 4.06QKK110 pKa = 9.88TILRR114 pKa = 11.84VCKK117 pKa = 9.73WNGRR121 pKa = 11.84YY122 pKa = 8.23MPSYY126 pKa = 10.52RR127 pKa = 11.84PTSTTNGYY135 pKa = 10.5DD136 pKa = 3.21CVINVNLNLRR146 pKa = 11.84FVDD149 pKa = 3.87QQSHH153 pKa = 5.85NDD155 pKa = 3.33VFGLSWSGDD164 pKa = 3.05RR165 pKa = 11.84LIIYY169 pKa = 10.05SVDD172 pKa = 3.21GVSSHH177 pKa = 6.6HH178 pKa = 6.76VDD180 pKa = 4.42GIGDD184 pKa = 4.25LDD186 pKa = 3.82VVSFRR191 pKa = 11.84CQLKK195 pKa = 7.94EE196 pKa = 4.08TCAHH200 pKa = 5.15QVVTKK205 pKa = 10.16PITAIVNTTSDD216 pKa = 3.69GFISGYY222 pKa = 7.31EE223 pKa = 3.86VCADD227 pKa = 3.28CDD229 pKa = 3.83GFPKK233 pKa = 10.72YY234 pKa = 10.73VFAVTDD240 pKa = 4.16GGAIPPNFQFDD251 pKa = 2.88NWFYY255 pKa = 10.18LTNSSTPISGMFTSVQPFNLSCVWPVPVLTSNSLPVHH292 pKa = 6.5FNMSLNNGAHH302 pKa = 6.45CNGYY306 pKa = 10.78SNLDD310 pKa = 3.5VGLVDD315 pKa = 5.3AMRR318 pKa = 11.84FSLNFTDD325 pKa = 3.65SQIIRR330 pKa = 11.84TGVISVITSTNVFNFSCTNSSVFGEE355 pKa = 4.38TNVIPFGDD363 pKa = 3.69VEE365 pKa = 4.23KK366 pKa = 9.67THH368 pKa = 6.5YY369 pKa = 10.86CFINYY374 pKa = 6.99FTNITTGEE382 pKa = 4.2SASQFVGILPPQVKK396 pKa = 9.71EE397 pKa = 3.69FVIMRR402 pKa = 11.84NGDD405 pKa = 3.34FHH407 pKa = 8.25LNGYY411 pKa = 10.09RR412 pKa = 11.84IFSVDD417 pKa = 3.14SVEE420 pKa = 4.69SVIFNISTKK429 pKa = 10.3DD430 pKa = 3.43GRR432 pKa = 11.84DD433 pKa = 3.2FWTVAFANNAEE444 pKa = 4.17VLTEE448 pKa = 4.01INATSIQNLLYY459 pKa = 10.82CNNPVNSIKK468 pKa = 10.55CQQLRR473 pKa = 11.84FNLDD477 pKa = 3.27DD478 pKa = 4.0GFYY481 pKa = 10.69SHH483 pKa = 7.19TFDD486 pKa = 5.35SVDD489 pKa = 3.28NLPRR493 pKa = 11.84TIVRR497 pKa = 11.84LPKK500 pKa = 10.35YY501 pKa = 8.13VTHH504 pKa = 6.86SIINVTVGINFEE516 pKa = 4.41NKK518 pKa = 8.8TLTNYY523 pKa = 10.83LIDD526 pKa = 4.39FAPDD530 pKa = 3.28DD531 pKa = 4.12VSIGKK536 pKa = 9.88DD537 pKa = 3.19NITTVCVDD545 pKa = 3.16TTTFTVRR552 pKa = 11.84LNVLAFSSFSTAVDD566 pKa = 3.57IQAGTCPFSYY576 pKa = 11.29NNLNNYY582 pKa = 9.99LGFGSLCFSTRR593 pKa = 11.84PNGGCMMSVVARR605 pKa = 11.84GYY607 pKa = 9.89FGEE610 pKa = 4.14FQKK613 pKa = 11.28VGVLYY618 pKa = 11.01VSFTKK623 pKa = 10.63GDD625 pKa = 3.57NVLGVPPNNVPTIGVSDD642 pKa = 3.7MSDD645 pKa = 2.83VKK647 pKa = 11.15LGVCTTFTIYY657 pKa = 10.65GHH659 pKa = 5.54TGRR662 pKa = 11.84GIINKK667 pKa = 9.73SNNTFVSGLFYY678 pKa = 10.84TSVTGNLLGFKK689 pKa = 10.45NSTTGEE695 pKa = 4.0IFSISPCQLTTQVAVISDD713 pKa = 4.37KK714 pKa = 11.0IVGVAAATTEE724 pKa = 4.1VKK726 pKa = 10.74LPFDD730 pKa = 4.24NYY732 pKa = 9.98VSLGSFYY739 pKa = 10.56YY740 pKa = 10.02HH741 pKa = 6.21YY742 pKa = 10.07RR743 pKa = 11.84NSSAEE748 pKa = 3.92LCKK751 pKa = 10.85NPSLMYY757 pKa = 10.57GGLGVCEE764 pKa = 5.02DD765 pKa = 3.57GRR767 pKa = 11.84LVNISRR773 pKa = 11.84SEE775 pKa = 4.02DD776 pKa = 3.45TFVPSAVISGNITIPANFSFVVQPEE801 pKa = 4.31YY802 pKa = 10.43IQIMTKK808 pKa = 9.87PVSVDD813 pKa = 2.99CSVYY817 pKa = 10.26VCNGNPRR824 pKa = 11.84CLQLLTQYY832 pKa = 11.57ASVCRR837 pKa = 11.84TVEE840 pKa = 4.29EE841 pKa = 4.13PLQLNARR848 pKa = 11.84LEE850 pKa = 4.16ALEE853 pKa = 4.37LTSMIIVQDD862 pKa = 3.48KK863 pKa = 8.43TLKK866 pKa = 10.86LGVASNFNDD875 pKa = 3.82TFDD878 pKa = 3.91LTLALPRR885 pKa = 11.84QHH887 pKa = 6.45QSRR890 pKa = 11.84SAIEE894 pKa = 4.54DD895 pKa = 3.65LLFSKK900 pKa = 10.43IVTSGLGTVDD910 pKa = 5.34DD911 pKa = 5.32DD912 pKa = 4.97YY913 pKa = 11.92KK914 pKa = 11.26EE915 pKa = 4.45CAAKK919 pKa = 9.16MANTIAEE926 pKa = 4.49AGCVQYY932 pKa = 11.5YY933 pKa = 9.78NGIMVLPGVVDD944 pKa = 4.33PSLLSQYY951 pKa = 9.05TAALTGAMVLGGVTAAAAIPFSLAVQARR979 pKa = 11.84LNYY982 pKa = 10.03LALQTDD988 pKa = 4.14VLQRR992 pKa = 11.84NQMILAQSFNAAMGNITVAFSGVSNAIQQTAQSLNTISQALNKK1035 pKa = 9.73VEE1037 pKa = 4.27NVVNEE1042 pKa = 3.91QGTALSQLTKK1052 pKa = 10.61QLASNFQAISSSIEE1066 pKa = 3.69DD1067 pKa = 3.97LYY1069 pKa = 11.84NRR1071 pKa = 11.84LDD1073 pKa = 3.6TLEE1076 pKa = 5.38ADD1078 pKa = 3.74QQVDD1082 pKa = 3.46RR1083 pKa = 11.84LITGRR1088 pKa = 11.84LAALNAFVTQQLLRR1102 pKa = 11.84YY1103 pKa = 8.9SEE1105 pKa = 4.28VRR1107 pKa = 11.84ASRR1110 pKa = 11.84QLAQEE1115 pKa = 4.75KK1116 pKa = 10.45INEE1119 pKa = 4.44CVKK1122 pKa = 10.53SQSSRR1127 pKa = 11.84YY1128 pKa = 8.35GFCGNGTHH1136 pKa = 5.87VFSVANAAPDD1146 pKa = 4.45GIMFLHH1152 pKa = 6.91ANLVPTAFIEE1162 pKa = 4.41VSAFAGVCVEE1172 pKa = 3.85GRR1174 pKa = 11.84ALVLRR1179 pKa = 11.84GRR1181 pKa = 11.84DD1182 pKa = 3.0EE1183 pKa = 4.97VLFQKK1188 pKa = 10.29PNTMQYY1194 pKa = 10.94LITPRR1199 pKa = 11.84LLFEE1203 pKa = 4.25PRR1205 pKa = 11.84VPVSADD1211 pKa = 3.25FVEE1214 pKa = 4.87VSSCNVTFINLTINEE1229 pKa = 4.53LPEE1232 pKa = 4.46LLPDD1236 pKa = 4.18YY1237 pKa = 10.98IDD1239 pKa = 3.45VNKK1242 pKa = 9.25TLKK1245 pKa = 10.34EE1246 pKa = 3.88FAATIPNRR1254 pKa = 11.84TEE1256 pKa = 3.59MQLTLNTYY1264 pKa = 9.78NATVLNLTDD1273 pKa = 3.74EE1274 pKa = 4.85VKK1276 pKa = 10.99SLMSQADD1283 pKa = 4.45DD1284 pKa = 3.54LTKK1287 pKa = 10.24IASEE1291 pKa = 4.28LNLTISKK1298 pKa = 10.27INNTLVEE1305 pKa = 4.51LEE1307 pKa = 3.89WLNRR1311 pKa = 11.84VEE1313 pKa = 5.97TYY1315 pKa = 10.4IKK1317 pKa = 9.3WPWYY1321 pKa = 7.96VWLAIAVTLIILVGLMLWCCLATGCCGCCSCLVNSCSDD1359 pKa = 3.33CGGRR1363 pKa = 11.84RR1364 pKa = 11.84LQRR1367 pKa = 11.84YY1368 pKa = 8.12EE1369 pKa = 3.64IEE1371 pKa = 4.3KK1372 pKa = 10.33VHH1374 pKa = 5.94IQQ1376 pKa = 3.19
Molecular weight: 150.93 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0U1WHB7|A0A0U1WHB7_9ALPC Spike glycoprotein OS=BtMf-AlphaCoV/JX2012 OX=1503288 PE=4 SV=1
MM1 pKa = 7.75ASVQFEE7 pKa = 4.47DD8 pKa = 3.64TNSRR12 pKa = 11.84GRR14 pKa = 11.84SGRR17 pKa = 11.84IPLSYY22 pKa = 9.62YY23 pKa = 10.63APVRR27 pKa = 11.84VADD30 pKa = 4.65DD31 pKa = 3.83KK32 pKa = 11.82LFLKK36 pKa = 10.66SMPGNGVPKK45 pKa = 10.76GMGGKK50 pKa = 9.77AEE52 pKa = 4.14QIGYY56 pKa = 6.34WTEE59 pKa = 3.6QRR61 pKa = 11.84RR62 pKa = 11.84WRR64 pKa = 11.84MNKK67 pKa = 8.32GQRR70 pKa = 11.84KK71 pKa = 8.92DD72 pKa = 4.4LPSNWHH78 pKa = 6.54FYY80 pKa = 10.83YY81 pKa = 10.82LGTGPHH87 pKa = 6.91ADD89 pKa = 2.92LKK91 pKa = 10.62FRR93 pKa = 11.84EE94 pKa = 4.21RR95 pKa = 11.84QVGVFWVAKK104 pKa = 9.45QGSKK108 pKa = 9.38TEE110 pKa = 4.03PTNLGTRR117 pKa = 11.84NRR119 pKa = 11.84NQQLRR124 pKa = 11.84IPEE127 pKa = 4.11FDD129 pKa = 3.1FTLPDD134 pKa = 3.28SLEE137 pKa = 4.22VVNSDD142 pKa = 2.99SRR144 pKa = 11.84PASRR148 pKa = 11.84SNSRR152 pKa = 11.84SQSKK156 pKa = 10.82SNNQSRR162 pKa = 11.84SNSKK166 pKa = 10.5ARR168 pKa = 11.84DD169 pKa = 3.42QSGTRR174 pKa = 11.84TPKK177 pKa = 10.4NASQNQKK184 pKa = 9.64PKK186 pKa = 10.77KK187 pKa = 9.04NQQEE191 pKa = 4.18DD192 pKa = 3.64LVAAVRR198 pKa = 11.84EE199 pKa = 4.15ALKK202 pKa = 11.01GLGIQQSRR210 pKa = 11.84SSSGKK215 pKa = 8.13STPARR220 pKa = 11.84SKK222 pKa = 11.13SPARR226 pKa = 11.84PSNKK230 pKa = 9.31KK231 pKa = 9.84QLDD234 pKa = 3.49KK235 pKa = 11.31PEE237 pKa = 4.08WKK239 pKa = 9.82RR240 pKa = 11.84VPNRR244 pKa = 11.84TEE246 pKa = 3.54NATTCFGQRR255 pKa = 11.84SVEE258 pKa = 4.2RR259 pKa = 11.84NCGSASVVGMGVEE272 pKa = 4.43APNFPQIAEE281 pKa = 4.98LIPTQAALFFGSRR294 pKa = 11.84VSTKK298 pKa = 10.23EE299 pKa = 3.51VGDD302 pKa = 3.76TVEE305 pKa = 4.23IKK307 pKa = 10.26YY308 pKa = 9.74HH309 pKa = 5.79YY310 pKa = 10.05KK311 pKa = 9.87MSVPKK316 pKa = 9.99TNPNLPYY323 pKa = 10.51FLQQVNAYY331 pKa = 9.7LDD333 pKa = 4.12PNSDD337 pKa = 3.17QPIPKK342 pKa = 9.52KK343 pKa = 10.36PKK345 pKa = 9.69AKK347 pKa = 10.09PEE349 pKa = 3.99EE350 pKa = 4.21VPQLNPAAPVFTPPAQEE367 pKa = 3.78PAVEE371 pKa = 4.41LEE373 pKa = 4.26MVDD376 pKa = 3.64EE377 pKa = 4.78VFDD380 pKa = 4.29IDD382 pKa = 4.1PLGDD386 pKa = 3.55SVAA389 pKa = 3.81
MM1 pKa = 7.75ASVQFEE7 pKa = 4.47DD8 pKa = 3.64TNSRR12 pKa = 11.84GRR14 pKa = 11.84SGRR17 pKa = 11.84IPLSYY22 pKa = 9.62YY23 pKa = 10.63APVRR27 pKa = 11.84VADD30 pKa = 4.65DD31 pKa = 3.83KK32 pKa = 11.82LFLKK36 pKa = 10.66SMPGNGVPKK45 pKa = 10.76GMGGKK50 pKa = 9.77AEE52 pKa = 4.14QIGYY56 pKa = 6.34WTEE59 pKa = 3.6QRR61 pKa = 11.84RR62 pKa = 11.84WRR64 pKa = 11.84MNKK67 pKa = 8.32GQRR70 pKa = 11.84KK71 pKa = 8.92DD72 pKa = 4.4LPSNWHH78 pKa = 6.54FYY80 pKa = 10.83YY81 pKa = 10.82LGTGPHH87 pKa = 6.91ADD89 pKa = 2.92LKK91 pKa = 10.62FRR93 pKa = 11.84EE94 pKa = 4.21RR95 pKa = 11.84QVGVFWVAKK104 pKa = 9.45QGSKK108 pKa = 9.38TEE110 pKa = 4.03PTNLGTRR117 pKa = 11.84NRR119 pKa = 11.84NQQLRR124 pKa = 11.84IPEE127 pKa = 4.11FDD129 pKa = 3.1FTLPDD134 pKa = 3.28SLEE137 pKa = 4.22VVNSDD142 pKa = 2.99SRR144 pKa = 11.84PASRR148 pKa = 11.84SNSRR152 pKa = 11.84SQSKK156 pKa = 10.82SNNQSRR162 pKa = 11.84SNSKK166 pKa = 10.5ARR168 pKa = 11.84DD169 pKa = 3.42QSGTRR174 pKa = 11.84TPKK177 pKa = 10.4NASQNQKK184 pKa = 9.64PKK186 pKa = 10.77KK187 pKa = 9.04NQQEE191 pKa = 4.18DD192 pKa = 3.64LVAAVRR198 pKa = 11.84EE199 pKa = 4.15ALKK202 pKa = 11.01GLGIQQSRR210 pKa = 11.84SSSGKK215 pKa = 8.13STPARR220 pKa = 11.84SKK222 pKa = 11.13SPARR226 pKa = 11.84PSNKK230 pKa = 9.31KK231 pKa = 9.84QLDD234 pKa = 3.49KK235 pKa = 11.31PEE237 pKa = 4.08WKK239 pKa = 9.82RR240 pKa = 11.84VPNRR244 pKa = 11.84TEE246 pKa = 3.54NATTCFGQRR255 pKa = 11.84SVEE258 pKa = 4.2RR259 pKa = 11.84NCGSASVVGMGVEE272 pKa = 4.43APNFPQIAEE281 pKa = 4.98LIPTQAALFFGSRR294 pKa = 11.84VSTKK298 pKa = 10.23EE299 pKa = 3.51VGDD302 pKa = 3.76TVEE305 pKa = 4.23IKK307 pKa = 10.26YY308 pKa = 9.74HH309 pKa = 5.79YY310 pKa = 10.05KK311 pKa = 9.87MSVPKK316 pKa = 9.99TNPNLPYY323 pKa = 10.51FLQQVNAYY331 pKa = 9.7LDD333 pKa = 4.12PNSDD337 pKa = 3.17QPIPKK342 pKa = 9.52KK343 pKa = 10.36PKK345 pKa = 9.69AKK347 pKa = 10.09PEE349 pKa = 3.99EE350 pKa = 4.21VPQLNPAAPVFTPPAQEE367 pKa = 3.78PAVEE371 pKa = 4.41LEE373 pKa = 4.26MVDD376 pKa = 3.64EE377 pKa = 4.78VFDD380 pKa = 4.29IDD382 pKa = 4.1PLGDD386 pKa = 3.55SVAA389 pKa = 3.81
Molecular weight: 43.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
9241 |
74 |
6931 |
1540.2 |
171.17 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.59 ± 0.216 | 3.344 ± 0.632 |
5.497 ± 0.792 | 4.123 ± 0.29 |
5.811 ± 0.288 | 6.471 ± 0.278 |
1.85 ± 0.256 | 5.378 ± 0.538 |
5.681 ± 1.041 | 8.224 ± 0.883 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.067 ± 0.198 | 6.298 ± 0.564 |
3.495 ± 0.76 | 2.77 ± 0.74 |
3.333 ± 0.553 | 7.478 ± 0.338 |
5.995 ± 0.675 | 10.042 ± 0.684 |
1.104 ± 0.277 | 4.448 ± 0.386 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
