
Wenzhou tombus-like virus 11
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.89
Get precalculated fractions of proteins
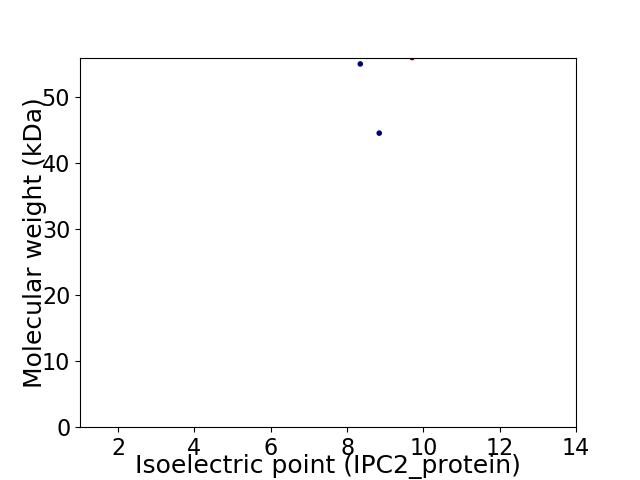
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KG22|A0A1L3KG22_9VIRU Uncharacterized protein OS=Wenzhou tombus-like virus 11 OX=1923664 PE=4 SV=1
MM1 pKa = 7.73KK2 pKa = 10.17GRR4 pKa = 11.84PISDD8 pKa = 3.31RR9 pKa = 11.84QLPGSSLRR17 pKa = 11.84TGEE20 pKa = 4.53CVHH23 pKa = 5.84SCRR26 pKa = 11.84RR27 pKa = 11.84YY28 pKa = 6.76TRR30 pKa = 11.84KK31 pKa = 9.17MFDD34 pKa = 3.12YY35 pKa = 9.18DD36 pKa = 3.46TDD38 pKa = 4.01FGGSDD43 pKa = 3.7SVVKK47 pKa = 8.86THH49 pKa = 7.1LSCVCNEE56 pKa = 3.63VLALKK61 pKa = 10.15NRR63 pKa = 11.84HH64 pKa = 5.37QLDD67 pKa = 3.46DD68 pKa = 3.69GARR71 pKa = 11.84YY72 pKa = 7.06TFKK75 pKa = 11.24GDD77 pKa = 3.01LTKK80 pKa = 10.1WVRR83 pKa = 11.84RR84 pKa = 11.84RR85 pKa = 11.84VKK87 pKa = 10.55PMTPWSEE94 pKa = 4.05DD95 pKa = 3.14MVIEE99 pKa = 4.53HH100 pKa = 7.77AIPSKK105 pKa = 10.84KK106 pKa = 9.74KK107 pKa = 10.12LLMSAKK113 pKa = 9.72EE114 pKa = 4.03SLKK117 pKa = 10.48QYY119 pKa = 10.95PLTEE123 pKa = 3.99KK124 pKa = 10.72DD125 pKa = 3.22AAIKK129 pKa = 10.18MFLKK133 pKa = 10.27DD134 pKa = 3.75DD135 pKa = 4.15KK136 pKa = 11.57YY137 pKa = 10.58HH138 pKa = 8.41GEE140 pKa = 4.16MKK142 pKa = 10.46DD143 pKa = 3.45PRR145 pKa = 11.84CIQYY149 pKa = 10.48RR150 pKa = 11.84NKK152 pKa = 10.47RR153 pKa = 11.84YY154 pKa = 9.5NLRR157 pKa = 11.84LGTYY161 pKa = 7.56LHH163 pKa = 6.77PLEE166 pKa = 5.0EE167 pKa = 4.74YY168 pKa = 10.78VMSWTHH174 pKa = 6.52RR175 pKa = 11.84GTHH178 pKa = 5.9IFAKK182 pKa = 9.05GRR184 pKa = 11.84NMRR187 pKa = 11.84QRR189 pKa = 11.84GRR191 pKa = 11.84DD192 pKa = 2.96IAAKK196 pKa = 10.13FNGIEE201 pKa = 4.09DD202 pKa = 4.31CVVLSIDD209 pKa = 3.37HH210 pKa = 6.77SKK212 pKa = 10.66FDD214 pKa = 3.75CHH216 pKa = 7.09VNRR219 pKa = 11.84QLLEE223 pKa = 4.23AEE225 pKa = 3.72HH226 pKa = 6.61WFYY229 pKa = 11.34NQCFGSEE236 pKa = 3.93EE237 pKa = 4.18LAFLLQLQIRR247 pKa = 11.84NKK249 pKa = 9.2GTTKK253 pKa = 10.64NGTRR257 pKa = 11.84YY258 pKa = 5.91TTINTRR264 pKa = 11.84MSGDD268 pKa = 3.38KK269 pKa = 9.66NTGLGNSLINYY280 pKa = 7.38MMIKK284 pKa = 9.4QVLMEE289 pKa = 4.32LAIKK293 pKa = 9.79HH294 pKa = 5.92NYY296 pKa = 9.27YY297 pKa = 10.04IDD299 pKa = 5.02GDD301 pKa = 3.96DD302 pKa = 3.88SNVFVQRR309 pKa = 11.84RR310 pKa = 11.84FAHH313 pKa = 5.83LVKK316 pKa = 10.81AEE318 pKa = 3.98LFSRR322 pKa = 11.84FGMVTKK328 pKa = 10.21IEE330 pKa = 4.12NVADD334 pKa = 4.27VIEE337 pKa = 5.63HH338 pKa = 7.37IDD340 pKa = 3.57FCQCRR345 pKa = 11.84PVFDD349 pKa = 3.98GSGYY353 pKa = 8.93TMVRR357 pKa = 11.84NPEE360 pKa = 3.78RR361 pKa = 11.84MLARR365 pKa = 11.84LPWVVGPIEE374 pKa = 4.16EE375 pKa = 4.65SRR377 pKa = 11.84ALDD380 pKa = 3.08ITYY383 pKa = 8.13ATGQCEE389 pKa = 3.91IAQGLGLPIGQYY401 pKa = 9.48IGQRR405 pKa = 11.84MCEE408 pKa = 3.98LGGKK412 pKa = 7.34MVRR415 pKa = 11.84LRR417 pKa = 11.84HH418 pKa = 5.49RR419 pKa = 11.84AWLEE423 pKa = 3.5KK424 pKa = 9.79MKK426 pKa = 10.25PGKK429 pKa = 9.5LQPIEE434 pKa = 3.78PAAGVRR440 pKa = 11.84EE441 pKa = 4.45SYY443 pKa = 11.19ALAWGLSVADD453 pKa = 3.82QLAIEE458 pKa = 4.44QTRR461 pKa = 11.84LVQPEE466 pKa = 4.02EE467 pKa = 4.79LYY469 pKa = 10.7TPGTLEE475 pKa = 3.86WLL477 pKa = 3.91
MM1 pKa = 7.73KK2 pKa = 10.17GRR4 pKa = 11.84PISDD8 pKa = 3.31RR9 pKa = 11.84QLPGSSLRR17 pKa = 11.84TGEE20 pKa = 4.53CVHH23 pKa = 5.84SCRR26 pKa = 11.84RR27 pKa = 11.84YY28 pKa = 6.76TRR30 pKa = 11.84KK31 pKa = 9.17MFDD34 pKa = 3.12YY35 pKa = 9.18DD36 pKa = 3.46TDD38 pKa = 4.01FGGSDD43 pKa = 3.7SVVKK47 pKa = 8.86THH49 pKa = 7.1LSCVCNEE56 pKa = 3.63VLALKK61 pKa = 10.15NRR63 pKa = 11.84HH64 pKa = 5.37QLDD67 pKa = 3.46DD68 pKa = 3.69GARR71 pKa = 11.84YY72 pKa = 7.06TFKK75 pKa = 11.24GDD77 pKa = 3.01LTKK80 pKa = 10.1WVRR83 pKa = 11.84RR84 pKa = 11.84RR85 pKa = 11.84VKK87 pKa = 10.55PMTPWSEE94 pKa = 4.05DD95 pKa = 3.14MVIEE99 pKa = 4.53HH100 pKa = 7.77AIPSKK105 pKa = 10.84KK106 pKa = 9.74KK107 pKa = 10.12LLMSAKK113 pKa = 9.72EE114 pKa = 4.03SLKK117 pKa = 10.48QYY119 pKa = 10.95PLTEE123 pKa = 3.99KK124 pKa = 10.72DD125 pKa = 3.22AAIKK129 pKa = 10.18MFLKK133 pKa = 10.27DD134 pKa = 3.75DD135 pKa = 4.15KK136 pKa = 11.57YY137 pKa = 10.58HH138 pKa = 8.41GEE140 pKa = 4.16MKK142 pKa = 10.46DD143 pKa = 3.45PRR145 pKa = 11.84CIQYY149 pKa = 10.48RR150 pKa = 11.84NKK152 pKa = 10.47RR153 pKa = 11.84YY154 pKa = 9.5NLRR157 pKa = 11.84LGTYY161 pKa = 7.56LHH163 pKa = 6.77PLEE166 pKa = 5.0EE167 pKa = 4.74YY168 pKa = 10.78VMSWTHH174 pKa = 6.52RR175 pKa = 11.84GTHH178 pKa = 5.9IFAKK182 pKa = 9.05GRR184 pKa = 11.84NMRR187 pKa = 11.84QRR189 pKa = 11.84GRR191 pKa = 11.84DD192 pKa = 2.96IAAKK196 pKa = 10.13FNGIEE201 pKa = 4.09DD202 pKa = 4.31CVVLSIDD209 pKa = 3.37HH210 pKa = 6.77SKK212 pKa = 10.66FDD214 pKa = 3.75CHH216 pKa = 7.09VNRR219 pKa = 11.84QLLEE223 pKa = 4.23AEE225 pKa = 3.72HH226 pKa = 6.61WFYY229 pKa = 11.34NQCFGSEE236 pKa = 3.93EE237 pKa = 4.18LAFLLQLQIRR247 pKa = 11.84NKK249 pKa = 9.2GTTKK253 pKa = 10.64NGTRR257 pKa = 11.84YY258 pKa = 5.91TTINTRR264 pKa = 11.84MSGDD268 pKa = 3.38KK269 pKa = 9.66NTGLGNSLINYY280 pKa = 7.38MMIKK284 pKa = 9.4QVLMEE289 pKa = 4.32LAIKK293 pKa = 9.79HH294 pKa = 5.92NYY296 pKa = 9.27YY297 pKa = 10.04IDD299 pKa = 5.02GDD301 pKa = 3.96DD302 pKa = 3.88SNVFVQRR309 pKa = 11.84RR310 pKa = 11.84FAHH313 pKa = 5.83LVKK316 pKa = 10.81AEE318 pKa = 3.98LFSRR322 pKa = 11.84FGMVTKK328 pKa = 10.21IEE330 pKa = 4.12NVADD334 pKa = 4.27VIEE337 pKa = 5.63HH338 pKa = 7.37IDD340 pKa = 3.57FCQCRR345 pKa = 11.84PVFDD349 pKa = 3.98GSGYY353 pKa = 8.93TMVRR357 pKa = 11.84NPEE360 pKa = 3.78RR361 pKa = 11.84MLARR365 pKa = 11.84LPWVVGPIEE374 pKa = 4.16EE375 pKa = 4.65SRR377 pKa = 11.84ALDD380 pKa = 3.08ITYY383 pKa = 8.13ATGQCEE389 pKa = 3.91IAQGLGLPIGQYY401 pKa = 9.48IGQRR405 pKa = 11.84MCEE408 pKa = 3.98LGGKK412 pKa = 7.34MVRR415 pKa = 11.84LRR417 pKa = 11.84HH418 pKa = 5.49RR419 pKa = 11.84AWLEE423 pKa = 3.5KK424 pKa = 9.79MKK426 pKa = 10.25PGKK429 pKa = 9.5LQPIEE434 pKa = 3.78PAAGVRR440 pKa = 11.84EE441 pKa = 4.45SYY443 pKa = 11.19ALAWGLSVADD453 pKa = 3.82QLAIEE458 pKa = 4.44QTRR461 pKa = 11.84LVQPEE466 pKa = 4.02EE467 pKa = 4.79LYY469 pKa = 10.7TPGTLEE475 pKa = 3.86WLL477 pKa = 3.91
Molecular weight: 54.99 kDa
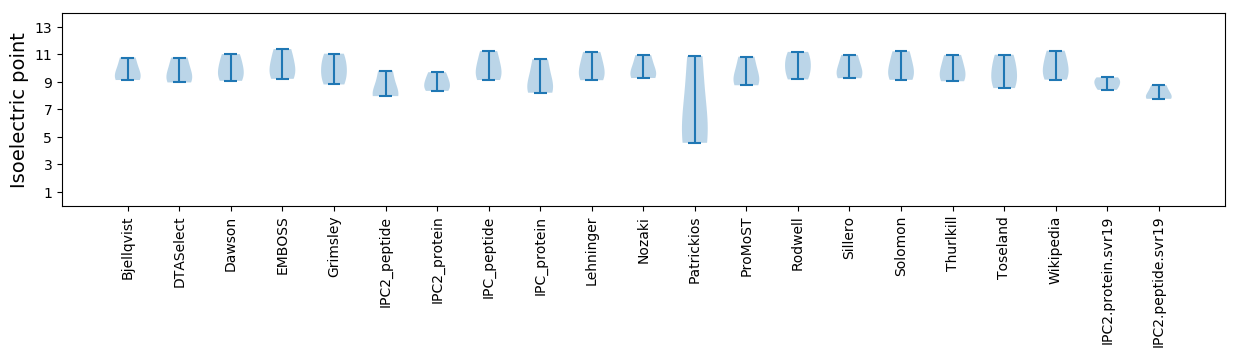
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KG58|A0A1L3KG58_9VIRU Uncharacterized protein OS=Wenzhou tombus-like virus 11 OX=1923664 PE=4 SV=1
MM1 pKa = 7.18SQPGSRR7 pKa = 11.84GQSRR11 pKa = 11.84GAGRR15 pKa = 11.84QTPNSGHH22 pKa = 7.17RR23 pKa = 11.84GRR25 pKa = 11.84DD26 pKa = 3.41GPRR29 pKa = 11.84RR30 pKa = 11.84PTGDD34 pKa = 3.48DD35 pKa = 3.06RR36 pKa = 11.84QPNQRR41 pKa = 11.84ARR43 pKa = 11.84GQHH46 pKa = 5.74PSGNARR52 pKa = 11.84GPQWQTVGRR61 pKa = 11.84RR62 pKa = 11.84PRR64 pKa = 11.84AANPEE69 pKa = 3.63ALMRR73 pKa = 11.84RR74 pKa = 11.84IHH76 pKa = 7.14ALEE79 pKa = 4.39QINKK83 pKa = 9.79DD84 pKa = 3.59LLIQLGRR91 pKa = 11.84INPGEE96 pKa = 4.21GSSGSTSRR104 pKa = 11.84GSTSGKK110 pKa = 8.58ATPEE114 pKa = 4.09KK115 pKa = 9.09PTPAPRR121 pKa = 11.84KK122 pKa = 9.56GPEE125 pKa = 4.01SRR127 pKa = 11.84NPTPVHH133 pKa = 5.46QASTATPRR141 pKa = 11.84GEE143 pKa = 4.08NQASRR148 pKa = 11.84TSKK151 pKa = 10.11GRR153 pKa = 11.84QRR155 pKa = 11.84CRR157 pKa = 11.84TLSNSHH163 pKa = 7.01LPRR166 pKa = 11.84VTTPNPFEE174 pKa = 4.12LLEE177 pKa = 4.19EE178 pKa = 4.44EE179 pKa = 5.03FPHH182 pKa = 7.13LSPGTLALAASVGSSGACTPTVKK205 pKa = 9.9TKK207 pKa = 10.59SGAPAGPSRR216 pKa = 11.84RR217 pKa = 11.84GRR219 pKa = 11.84SPPSTQRR226 pKa = 11.84ASSLPALNAQHH237 pKa = 6.9RR238 pKa = 11.84APRR241 pKa = 11.84AGGQQPSRR249 pKa = 11.84TFVTSSTPTQGTSSQRR265 pKa = 11.84PGATAEE271 pKa = 4.25PKK273 pKa = 9.91VAPNQAGSDD282 pKa = 3.8PAPQPRR288 pKa = 11.84VRR290 pKa = 11.84FRR292 pKa = 11.84PGDD295 pKa = 3.67FGWRR299 pKa = 11.84GAAQHH304 pKa = 6.37GVFHH308 pKa = 7.05SPTVASPRR316 pKa = 11.84YY317 pKa = 9.93AGACLEE323 pKa = 4.3ASNKK327 pKa = 9.76KK328 pKa = 10.55GPLANDD334 pKa = 3.58SNPVFILKK342 pKa = 10.3PEE344 pKa = 3.7VRR346 pKa = 11.84TDD348 pKa = 3.4VEE350 pKa = 4.58CGVGSTRR357 pKa = 11.84ANLARR362 pKa = 11.84LLDD365 pKa = 4.36LKK367 pKa = 11.08EE368 pKa = 4.0MSLDD372 pKa = 3.79SLYY375 pKa = 10.67QDD377 pKa = 4.46SGLDD381 pKa = 3.54PNSWKK386 pKa = 10.75DD387 pKa = 3.58LVSFTIDD394 pKa = 2.81KK395 pKa = 10.98AKK397 pKa = 10.9ALVVDD402 pKa = 3.7EE403 pKa = 4.29FMYY406 pKa = 11.15YY407 pKa = 10.45EE408 pKa = 4.21LVSRR412 pKa = 11.84FPFTVRR418 pKa = 11.84TADD421 pKa = 3.85LARR424 pKa = 11.84KK425 pKa = 7.79MFNHH429 pKa = 6.69LNILMKK435 pKa = 10.77QFDD438 pKa = 3.93TRR440 pKa = 11.84MYY442 pKa = 9.2TAKK445 pKa = 9.88QLYY448 pKa = 9.19RR449 pKa = 11.84IKK451 pKa = 10.54QATVRR456 pKa = 11.84AALLPPAEE464 pKa = 4.24EE465 pKa = 4.01LLTRR469 pKa = 11.84KK470 pKa = 10.23LIQQEE475 pKa = 4.17SKK477 pKa = 11.0EE478 pKa = 4.11MKK480 pKa = 9.87KK481 pKa = 10.96YY482 pKa = 10.7NAFAEE487 pKa = 4.31KK488 pKa = 10.59GNAGSVFAPGAGIVAAATSLAQTRR512 pKa = 11.84VGLSPDD518 pKa = 3.05KK519 pKa = 10.89KK520 pKa = 10.76
MM1 pKa = 7.18SQPGSRR7 pKa = 11.84GQSRR11 pKa = 11.84GAGRR15 pKa = 11.84QTPNSGHH22 pKa = 7.17RR23 pKa = 11.84GRR25 pKa = 11.84DD26 pKa = 3.41GPRR29 pKa = 11.84RR30 pKa = 11.84PTGDD34 pKa = 3.48DD35 pKa = 3.06RR36 pKa = 11.84QPNQRR41 pKa = 11.84ARR43 pKa = 11.84GQHH46 pKa = 5.74PSGNARR52 pKa = 11.84GPQWQTVGRR61 pKa = 11.84RR62 pKa = 11.84PRR64 pKa = 11.84AANPEE69 pKa = 3.63ALMRR73 pKa = 11.84RR74 pKa = 11.84IHH76 pKa = 7.14ALEE79 pKa = 4.39QINKK83 pKa = 9.79DD84 pKa = 3.59LLIQLGRR91 pKa = 11.84INPGEE96 pKa = 4.21GSSGSTSRR104 pKa = 11.84GSTSGKK110 pKa = 8.58ATPEE114 pKa = 4.09KK115 pKa = 9.09PTPAPRR121 pKa = 11.84KK122 pKa = 9.56GPEE125 pKa = 4.01SRR127 pKa = 11.84NPTPVHH133 pKa = 5.46QASTATPRR141 pKa = 11.84GEE143 pKa = 4.08NQASRR148 pKa = 11.84TSKK151 pKa = 10.11GRR153 pKa = 11.84QRR155 pKa = 11.84CRR157 pKa = 11.84TLSNSHH163 pKa = 7.01LPRR166 pKa = 11.84VTTPNPFEE174 pKa = 4.12LLEE177 pKa = 4.19EE178 pKa = 4.44EE179 pKa = 5.03FPHH182 pKa = 7.13LSPGTLALAASVGSSGACTPTVKK205 pKa = 9.9TKK207 pKa = 10.59SGAPAGPSRR216 pKa = 11.84RR217 pKa = 11.84GRR219 pKa = 11.84SPPSTQRR226 pKa = 11.84ASSLPALNAQHH237 pKa = 6.9RR238 pKa = 11.84APRR241 pKa = 11.84AGGQQPSRR249 pKa = 11.84TFVTSSTPTQGTSSQRR265 pKa = 11.84PGATAEE271 pKa = 4.25PKK273 pKa = 9.91VAPNQAGSDD282 pKa = 3.8PAPQPRR288 pKa = 11.84VRR290 pKa = 11.84FRR292 pKa = 11.84PGDD295 pKa = 3.67FGWRR299 pKa = 11.84GAAQHH304 pKa = 6.37GVFHH308 pKa = 7.05SPTVASPRR316 pKa = 11.84YY317 pKa = 9.93AGACLEE323 pKa = 4.3ASNKK327 pKa = 9.76KK328 pKa = 10.55GPLANDD334 pKa = 3.58SNPVFILKK342 pKa = 10.3PEE344 pKa = 3.7VRR346 pKa = 11.84TDD348 pKa = 3.4VEE350 pKa = 4.58CGVGSTRR357 pKa = 11.84ANLARR362 pKa = 11.84LLDD365 pKa = 4.36LKK367 pKa = 11.08EE368 pKa = 4.0MSLDD372 pKa = 3.79SLYY375 pKa = 10.67QDD377 pKa = 4.46SGLDD381 pKa = 3.54PNSWKK386 pKa = 10.75DD387 pKa = 3.58LVSFTIDD394 pKa = 2.81KK395 pKa = 10.98AKK397 pKa = 10.9ALVVDD402 pKa = 3.7EE403 pKa = 4.29FMYY406 pKa = 11.15YY407 pKa = 10.45EE408 pKa = 4.21LVSRR412 pKa = 11.84FPFTVRR418 pKa = 11.84TADD421 pKa = 3.85LARR424 pKa = 11.84KK425 pKa = 7.79MFNHH429 pKa = 6.69LNILMKK435 pKa = 10.77QFDD438 pKa = 3.93TRR440 pKa = 11.84MYY442 pKa = 9.2TAKK445 pKa = 9.88QLYY448 pKa = 9.19RR449 pKa = 11.84IKK451 pKa = 10.54QATVRR456 pKa = 11.84AALLPPAEE464 pKa = 4.24EE465 pKa = 4.01LLTRR469 pKa = 11.84KK470 pKa = 10.23LIQQEE475 pKa = 4.17SKK477 pKa = 11.0EE478 pKa = 4.11MKK480 pKa = 9.87KK481 pKa = 10.96YY482 pKa = 10.7NAFAEE487 pKa = 4.31KK488 pKa = 10.59GNAGSVFAPGAGIVAAATSLAQTRR512 pKa = 11.84VGLSPDD518 pKa = 3.05KK519 pKa = 10.89KK520 pKa = 10.76
Molecular weight: 55.95 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1399 |
402 |
520 |
466.3 |
51.82 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.72 ± 1.368 | 1.43 ± 0.467 |
4.718 ± 0.572 | 4.861 ± 0.591 |
3.717 ± 0.494 | 7.863 ± 0.445 |
2.073 ± 0.456 | 3.574 ± 0.858 |
6.004 ± 0.468 | 8.149 ± 0.561 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.716 ± 0.63 | 3.788 ± 0.12 |
6.505 ± 1.534 | 5.075 ± 0.375 |
8.077 ± 0.735 | 7.577 ± 1.181 |
6.934 ± 0.728 | 5.575 ± 0.55 |
1.072 ± 0.284 | 2.573 ± 0.68 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
