
Wuhan aphid virus 2
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified ssRNA viruses; unclassified ssRNA positive-strand viruses
Average proteome isoelectric point is 7.14
Get precalculated fractions of proteins
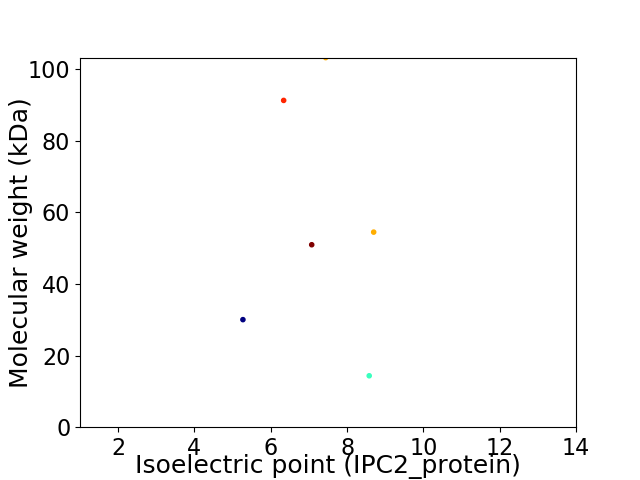
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0P0QKC6|A0A0P0QKC6_9VIRU Uncharacterized protein OS=Wuhan aphid virus 2 OX=1746068 GN=VP3 PE=4 SV=1
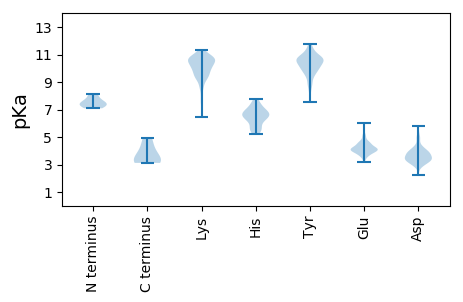
MM1 pKa = 7.71ANLIKK6 pKa = 10.4YY7 pKa = 9.81VIMLRR12 pKa = 11.84VVAAFAGTGMDD23 pKa = 4.76FMEE26 pKa = 5.64KK27 pKa = 10.19DD28 pKa = 3.98GKK30 pKa = 9.28TKK32 pKa = 10.63DD33 pKa = 3.3CAIYY37 pKa = 10.74DD38 pKa = 4.07EE39 pKa = 4.41AWRR42 pKa = 11.84VAKK45 pKa = 10.22CASSLRR51 pKa = 11.84EE52 pKa = 3.71GNMISNKK59 pKa = 9.8LFSFYY64 pKa = 10.9LQVALRR70 pKa = 11.84KK71 pKa = 9.5VPSSSAYY78 pKa = 8.64TSIGFTTGVDD88 pKa = 3.04RR89 pKa = 11.84EE90 pKa = 4.54LIVGKK95 pKa = 9.92QSHH98 pKa = 6.48YY99 pKa = 11.6DD100 pKa = 3.41FDD102 pKa = 3.27QWLYY106 pKa = 9.94TLTYY110 pKa = 10.44DD111 pKa = 4.24PRR113 pKa = 11.84TMLRR117 pKa = 11.84KK118 pKa = 9.02VAVISLHH125 pKa = 6.47RR126 pKa = 11.84EE127 pKa = 3.52YY128 pKa = 10.58TRR130 pKa = 11.84PVLFFSKK137 pKa = 9.38TVPEE141 pKa = 4.6EE142 pKa = 4.0LPVWDD147 pKa = 4.88DD148 pKa = 3.9LMVPSLRR155 pKa = 11.84LAGSGRR161 pKa = 11.84EE162 pKa = 3.73FFIYY166 pKa = 9.89KK167 pKa = 10.12DD168 pKa = 3.54KK169 pKa = 11.05NASPFSPSNAEE180 pKa = 4.02SCSAFYY186 pKa = 10.59DD187 pKa = 3.5WEE189 pKa = 4.16NRR191 pKa = 11.84EE192 pKa = 4.17SFEE195 pKa = 4.52DD196 pKa = 3.62ALGSVMIMDD205 pKa = 4.21PRR207 pKa = 11.84LWCPAMEE214 pKa = 4.68SNRR217 pKa = 11.84FDD219 pKa = 3.4IVVGDD224 pKa = 3.9LEE226 pKa = 4.59EE227 pKa = 4.5IKK229 pKa = 8.25TTSLDD234 pKa = 3.49SKK236 pKa = 10.7LQYY239 pKa = 10.27QSIWEE244 pKa = 4.17VTKK247 pKa = 10.79KK248 pKa = 9.48GTEE251 pKa = 3.86GGRR254 pKa = 11.84LDD256 pKa = 4.05EE257 pKa = 5.07NYY259 pKa = 10.86LSVFF263 pKa = 3.65
MM1 pKa = 7.71ANLIKK6 pKa = 10.4YY7 pKa = 9.81VIMLRR12 pKa = 11.84VVAAFAGTGMDD23 pKa = 4.76FMEE26 pKa = 5.64KK27 pKa = 10.19DD28 pKa = 3.98GKK30 pKa = 9.28TKK32 pKa = 10.63DD33 pKa = 3.3CAIYY37 pKa = 10.74DD38 pKa = 4.07EE39 pKa = 4.41AWRR42 pKa = 11.84VAKK45 pKa = 10.22CASSLRR51 pKa = 11.84EE52 pKa = 3.71GNMISNKK59 pKa = 9.8LFSFYY64 pKa = 10.9LQVALRR70 pKa = 11.84KK71 pKa = 9.5VPSSSAYY78 pKa = 8.64TSIGFTTGVDD88 pKa = 3.04RR89 pKa = 11.84EE90 pKa = 4.54LIVGKK95 pKa = 9.92QSHH98 pKa = 6.48YY99 pKa = 11.6DD100 pKa = 3.41FDD102 pKa = 3.27QWLYY106 pKa = 9.94TLTYY110 pKa = 10.44DD111 pKa = 4.24PRR113 pKa = 11.84TMLRR117 pKa = 11.84KK118 pKa = 9.02VAVISLHH125 pKa = 6.47RR126 pKa = 11.84EE127 pKa = 3.52YY128 pKa = 10.58TRR130 pKa = 11.84PVLFFSKK137 pKa = 9.38TVPEE141 pKa = 4.6EE142 pKa = 4.0LPVWDD147 pKa = 4.88DD148 pKa = 3.9LMVPSLRR155 pKa = 11.84LAGSGRR161 pKa = 11.84EE162 pKa = 3.73FFIYY166 pKa = 9.89KK167 pKa = 10.12DD168 pKa = 3.54KK169 pKa = 11.05NASPFSPSNAEE180 pKa = 4.02SCSAFYY186 pKa = 10.59DD187 pKa = 3.5WEE189 pKa = 4.16NRR191 pKa = 11.84EE192 pKa = 4.17SFEE195 pKa = 4.52DD196 pKa = 3.62ALGSVMIMDD205 pKa = 4.21PRR207 pKa = 11.84LWCPAMEE214 pKa = 4.68SNRR217 pKa = 11.84FDD219 pKa = 3.4IVVGDD224 pKa = 3.9LEE226 pKa = 4.59EE227 pKa = 4.5IKK229 pKa = 8.25TTSLDD234 pKa = 3.49SKK236 pKa = 10.7LQYY239 pKa = 10.27QSIWEE244 pKa = 4.17VTKK247 pKa = 10.79KK248 pKa = 9.48GTEE251 pKa = 3.86GGRR254 pKa = 11.84LDD256 pKa = 4.05EE257 pKa = 5.07NYY259 pKa = 10.86LSVFF263 pKa = 3.65
Molecular weight: 30.06 kDa
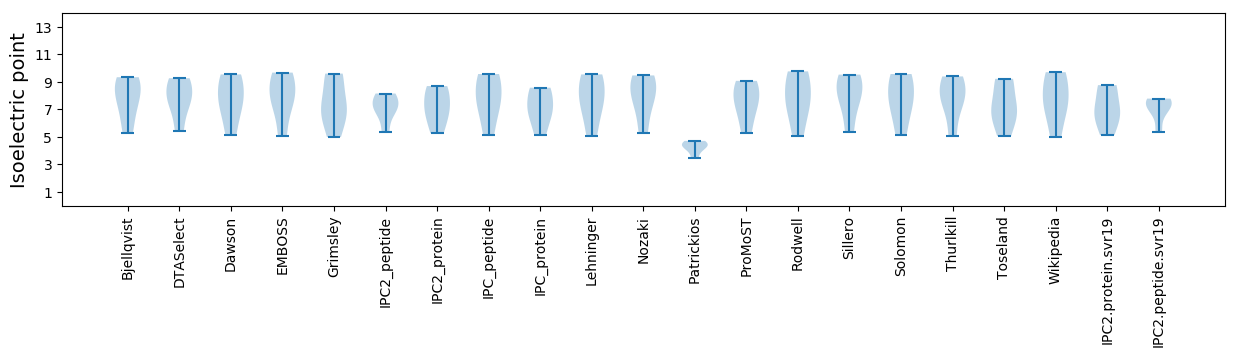
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0P0QKF2|A0A0P0QKF2_9VIRU Putative glycoprotein OS=Wuhan aphid virus 2 OX=1746068 GN=VP1 PE=4 SV=1
MM1 pKa = 8.13DD2 pKa = 3.64MTKK5 pKa = 10.66KK6 pKa = 10.62DD7 pKa = 3.41MEE9 pKa = 4.44RR10 pKa = 11.84AQEE13 pKa = 3.76RR14 pKa = 11.84AKK16 pKa = 10.59ILEE19 pKa = 4.14KK20 pKa = 10.58DD21 pKa = 3.8RR22 pKa = 11.84KK23 pKa = 10.03RR24 pKa = 11.84KK25 pKa = 8.87MKK27 pKa = 10.17EE28 pKa = 3.65FQEE31 pKa = 4.25SQKK34 pKa = 10.48MKK36 pKa = 10.49GKK38 pKa = 10.07FKK40 pKa = 10.65GRR42 pKa = 11.84VRR44 pKa = 11.84FWFDD48 pKa = 2.63EE49 pKa = 3.89RR50 pKa = 11.84SYY52 pKa = 11.73VVVRR56 pKa = 11.84DD57 pKa = 3.74ILLSPVFNLVGDD69 pKa = 4.24KK70 pKa = 11.04AHH72 pKa = 6.1MMLVGGYY79 pKa = 9.22LGAVLGVAFLAGRR92 pKa = 11.84IWGWLVPMLYY102 pKa = 9.52MGLIYY107 pKa = 10.5SDD109 pKa = 3.81DD110 pKa = 3.71TNAYY114 pKa = 8.75IAKK117 pKa = 9.71CLLTIWLVSKK127 pKa = 9.71VVKK130 pKa = 10.53CVFKK134 pKa = 10.93KK135 pKa = 10.92KK136 pKa = 10.03EE137 pKa = 4.08DD138 pKa = 4.03TYY140 pKa = 10.9HH141 pKa = 6.2HH142 pKa = 6.73AMMNVYY148 pKa = 10.08HH149 pKa = 7.31KK150 pKa = 9.26PTDD153 pKa = 2.82WMAIGNGIGFYY164 pKa = 10.67VEE166 pKa = 3.51IGIIIGLTYY175 pKa = 10.63LVYY178 pKa = 10.97GDD180 pKa = 4.54GYY182 pKa = 9.7GHH184 pKa = 7.62AFHH187 pKa = 6.66GVWVITNIMILSRR200 pKa = 11.84RR201 pKa = 11.84IPGVGGNSNLGLITLMCLVVIVVALLPEE229 pKa = 4.48VYY231 pKa = 10.22GVLLRR236 pKa = 11.84TTVKK240 pKa = 9.4TLEE243 pKa = 4.29PKK245 pKa = 10.55RR246 pKa = 11.84MAEE249 pKa = 4.04EE250 pKa = 5.14GIGMAAPIGHH260 pKa = 7.56LAGSFSRR267 pKa = 11.84PMDD270 pKa = 3.89YY271 pKa = 10.7IFGLSTTSFLDD282 pKa = 3.36VVRR285 pKa = 11.84ASCGNFLTGWLIFDD299 pKa = 3.87DD300 pKa = 4.03WFGAGHH306 pKa = 7.76ALTTATIIKK315 pKa = 9.29MKK317 pKa = 9.94GKK319 pKa = 9.64EE320 pKa = 3.9VTVPGSAGLYY330 pKa = 9.87GGLGAFWIAGDD341 pKa = 3.78IFLSATSGAVLRR353 pKa = 11.84VIVTVMSIAFSFWMWNWTGSKK374 pKa = 9.15VWSGRR379 pKa = 11.84GKK381 pKa = 10.91GVMLTEE387 pKa = 3.89ARR389 pKa = 11.84ANFGFVFGNGPVGMRR404 pKa = 11.84IAVLRR409 pKa = 11.84ACLFVATMCYY419 pKa = 10.55ALKK422 pKa = 10.55SDD424 pKa = 3.92GNIATGLILMMTITMGSEE442 pKa = 3.46RR443 pKa = 11.84AMTIMLGCMTMHH455 pKa = 7.45LSLFFQGIGKK465 pKa = 9.12NKK467 pKa = 9.93PITEE471 pKa = 4.3SLRR474 pKa = 11.84EE475 pKa = 4.18STVDD479 pKa = 3.27AYY481 pKa = 11.54SHH483 pKa = 7.03FDD485 pKa = 3.44GNASAGTEE493 pKa = 4.02PGG495 pKa = 3.23
MM1 pKa = 8.13DD2 pKa = 3.64MTKK5 pKa = 10.66KK6 pKa = 10.62DD7 pKa = 3.41MEE9 pKa = 4.44RR10 pKa = 11.84AQEE13 pKa = 3.76RR14 pKa = 11.84AKK16 pKa = 10.59ILEE19 pKa = 4.14KK20 pKa = 10.58DD21 pKa = 3.8RR22 pKa = 11.84KK23 pKa = 10.03RR24 pKa = 11.84KK25 pKa = 8.87MKK27 pKa = 10.17EE28 pKa = 3.65FQEE31 pKa = 4.25SQKK34 pKa = 10.48MKK36 pKa = 10.49GKK38 pKa = 10.07FKK40 pKa = 10.65GRR42 pKa = 11.84VRR44 pKa = 11.84FWFDD48 pKa = 2.63EE49 pKa = 3.89RR50 pKa = 11.84SYY52 pKa = 11.73VVVRR56 pKa = 11.84DD57 pKa = 3.74ILLSPVFNLVGDD69 pKa = 4.24KK70 pKa = 11.04AHH72 pKa = 6.1MMLVGGYY79 pKa = 9.22LGAVLGVAFLAGRR92 pKa = 11.84IWGWLVPMLYY102 pKa = 9.52MGLIYY107 pKa = 10.5SDD109 pKa = 3.81DD110 pKa = 3.71TNAYY114 pKa = 8.75IAKK117 pKa = 9.71CLLTIWLVSKK127 pKa = 9.71VVKK130 pKa = 10.53CVFKK134 pKa = 10.93KK135 pKa = 10.92KK136 pKa = 10.03EE137 pKa = 4.08DD138 pKa = 4.03TYY140 pKa = 10.9HH141 pKa = 6.2HH142 pKa = 6.73AMMNVYY148 pKa = 10.08HH149 pKa = 7.31KK150 pKa = 9.26PTDD153 pKa = 2.82WMAIGNGIGFYY164 pKa = 10.67VEE166 pKa = 3.51IGIIIGLTYY175 pKa = 10.63LVYY178 pKa = 10.97GDD180 pKa = 4.54GYY182 pKa = 9.7GHH184 pKa = 7.62AFHH187 pKa = 6.66GVWVITNIMILSRR200 pKa = 11.84RR201 pKa = 11.84IPGVGGNSNLGLITLMCLVVIVVALLPEE229 pKa = 4.48VYY231 pKa = 10.22GVLLRR236 pKa = 11.84TTVKK240 pKa = 9.4TLEE243 pKa = 4.29PKK245 pKa = 10.55RR246 pKa = 11.84MAEE249 pKa = 4.04EE250 pKa = 5.14GIGMAAPIGHH260 pKa = 7.56LAGSFSRR267 pKa = 11.84PMDD270 pKa = 3.89YY271 pKa = 10.7IFGLSTTSFLDD282 pKa = 3.36VVRR285 pKa = 11.84ASCGNFLTGWLIFDD299 pKa = 3.87DD300 pKa = 4.03WFGAGHH306 pKa = 7.76ALTTATIIKK315 pKa = 9.29MKK317 pKa = 9.94GKK319 pKa = 9.64EE320 pKa = 3.9VTVPGSAGLYY330 pKa = 9.87GGLGAFWIAGDD341 pKa = 3.78IFLSATSGAVLRR353 pKa = 11.84VIVTVMSIAFSFWMWNWTGSKK374 pKa = 9.15VWSGRR379 pKa = 11.84GKK381 pKa = 10.91GVMLTEE387 pKa = 3.89ARR389 pKa = 11.84ANFGFVFGNGPVGMRR404 pKa = 11.84IAVLRR409 pKa = 11.84ACLFVATMCYY419 pKa = 10.55ALKK422 pKa = 10.55SDD424 pKa = 3.92GNIATGLILMMTITMGSEE442 pKa = 3.46RR443 pKa = 11.84AMTIMLGCMTMHH455 pKa = 7.45LSLFFQGIGKK465 pKa = 9.12NKK467 pKa = 9.93PITEE471 pKa = 4.3SLRR474 pKa = 11.84EE475 pKa = 4.18STVDD479 pKa = 3.27AYY481 pKa = 11.54SHH483 pKa = 7.03FDD485 pKa = 3.44GNASAGTEE493 pKa = 4.02PGG495 pKa = 3.23
Molecular weight: 54.51 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3062 |
131 |
907 |
510.3 |
57.42 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.336 ± 0.59 | 1.47 ± 0.17 |
5.389 ± 0.355 | 5.585 ± 0.752 |
4.311 ± 0.438 | 8.165 ± 0.692 |
2.09 ± 0.257 | 5.944 ± 0.404 |
6.434 ± 0.249 | 9.471 ± 0.329 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.625 ± 0.548 | 3.527 ± 0.291 |
3.821 ± 0.295 | 1.96 ± 0.247 |
5.127 ± 0.502 | 6.434 ± 0.851 |
6.434 ± 0.343 | 7.479 ± 0.294 |
2.221 ± 0.246 | 4.18 ± 0.348 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
