
Torque teno virus 28
Taxonomy: Viruses; Anelloviridae; Alphatorquevirus
Average proteome isoelectric point is 7.85
Get precalculated fractions of proteins
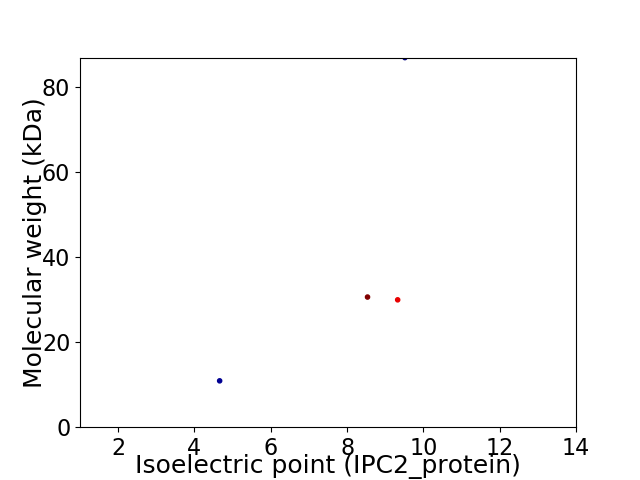
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
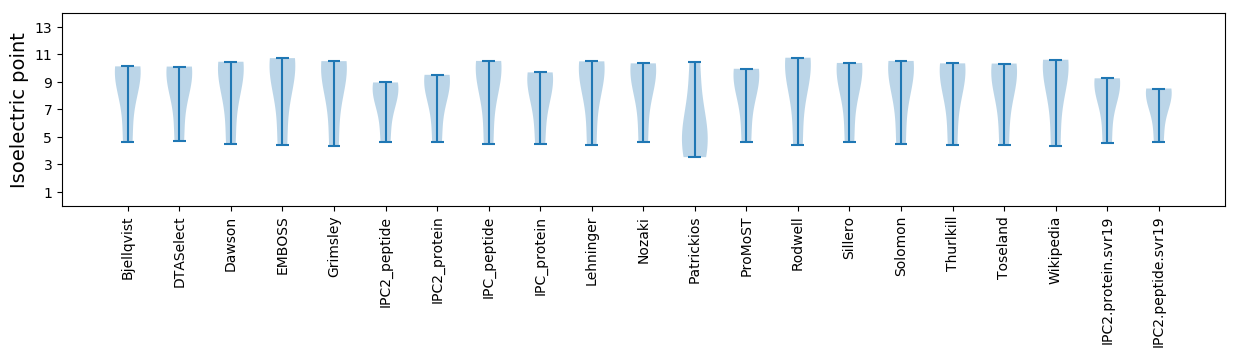
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q8V7I1|Q8V7I1_9VIRU Capsid protein OS=Torque teno virus 28 OX=687367 PE=3 SV=1
MM1 pKa = 7.33SWRR4 pKa = 11.84PPAYY8 pKa = 10.19NLAQRR13 pKa = 11.84EE14 pKa = 4.27EE15 pKa = 4.03HH16 pKa = 6.36WYY18 pKa = 10.05KK19 pKa = 11.02AILEE23 pKa = 4.29SHH25 pKa = 5.85STFCGCSDD33 pKa = 3.83VVRR36 pKa = 11.84HH37 pKa = 5.51FCVLASRR44 pKa = 11.84LATNPSVIPALPAPPEE60 pKa = 3.94QQPRR64 pKa = 11.84PGGDD68 pKa = 3.09TEE70 pKa = 4.61GAPGDD75 pKa = 4.46PGDD78 pKa = 4.48AGAGRR83 pKa = 11.84YY84 pKa = 9.19AEE86 pKa = 4.5EE87 pKa = 4.4DD88 pKa = 3.66LEE90 pKa = 4.37EE91 pKa = 4.54LFAAAAKK98 pKa = 10.76DD99 pKa = 3.78DD100 pKa = 3.79MM101 pKa = 6.43
MM1 pKa = 7.33SWRR4 pKa = 11.84PPAYY8 pKa = 10.19NLAQRR13 pKa = 11.84EE14 pKa = 4.27EE15 pKa = 4.03HH16 pKa = 6.36WYY18 pKa = 10.05KK19 pKa = 11.02AILEE23 pKa = 4.29SHH25 pKa = 5.85STFCGCSDD33 pKa = 3.83VVRR36 pKa = 11.84HH37 pKa = 5.51FCVLASRR44 pKa = 11.84LATNPSVIPALPAPPEE60 pKa = 3.94QQPRR64 pKa = 11.84PGGDD68 pKa = 3.09TEE70 pKa = 4.61GAPGDD75 pKa = 4.46PGDD78 pKa = 4.48AGAGRR83 pKa = 11.84YY84 pKa = 9.19AEE86 pKa = 4.5EE87 pKa = 4.4DD88 pKa = 3.66LEE90 pKa = 4.37EE91 pKa = 4.54LFAAAAKK98 pKa = 10.76DD99 pKa = 3.78DD100 pKa = 3.79MM101 pKa = 6.43
Molecular weight: 10.91 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q8V7I3|Q8V7I3_9VIRU Uncharacterized protein OS=Torque teno virus 28 OX=687367 PE=4 SV=1
MM1 pKa = 7.33SWRR4 pKa = 11.84PPAYY8 pKa = 10.19NLAQRR13 pKa = 11.84EE14 pKa = 4.27EE15 pKa = 4.03HH16 pKa = 6.36WYY18 pKa = 10.05KK19 pKa = 11.02AILEE23 pKa = 4.29SHH25 pKa = 5.85STFCGCSDD33 pKa = 3.83VVRR36 pKa = 11.84HH37 pKa = 5.51FCVLASRR44 pKa = 11.84LATNPSVIPALPAPPEE60 pKa = 3.94QQPRR64 pKa = 11.84PGGDD68 pKa = 3.09TEE70 pKa = 4.61GAPGDD75 pKa = 4.46PGDD78 pKa = 4.48AGAGRR83 pKa = 11.84YY84 pKa = 9.19AEE86 pKa = 4.5EE87 pKa = 4.4DD88 pKa = 3.66LEE90 pKa = 4.37EE91 pKa = 4.54LFAAAAKK98 pKa = 10.66DD99 pKa = 3.58DD100 pKa = 3.62MRR102 pKa = 11.84RR103 pKa = 11.84VQKK106 pKa = 9.49TPNGNPVRR114 pKa = 11.84GSHH117 pKa = 6.1RR118 pKa = 11.84KK119 pKa = 7.58PRR121 pKa = 11.84RR122 pKa = 11.84RR123 pKa = 11.84RR124 pKa = 11.84LLFTQSTPKK133 pKa = 10.18RR134 pKa = 11.84ARR136 pKa = 11.84EE137 pKa = 3.75QPVRR141 pKa = 11.84RR142 pKa = 11.84RR143 pKa = 11.84DD144 pKa = 3.2SPRR147 pKa = 11.84RR148 pKa = 11.84GDD150 pKa = 3.53PKK152 pKa = 10.57TATTQAAQPAATAAASPQRR171 pKa = 11.84GTQTSPGRR179 pKa = 11.84RR180 pKa = 11.84PPTPEE185 pKa = 4.17RR186 pKa = 11.84SPLGPPPIIIQGDD199 pKa = 4.68PIPDD203 pKa = 3.78LLFPSTGKK211 pKa = 9.8KK212 pKa = 9.82RR213 pKa = 11.84KK214 pKa = 8.94FSKK217 pKa = 10.58FDD219 pKa = 3.19WEE221 pKa = 4.34TEE223 pKa = 3.97AEE225 pKa = 3.77IGRR228 pKa = 11.84WLRR231 pKa = 11.84RR232 pKa = 11.84PMRR235 pKa = 11.84FYY237 pKa = 10.66PSDD240 pKa = 3.84PPHH243 pKa = 6.43YY244 pKa = 9.4PWLPPKK250 pKa = 10.23RR251 pKa = 11.84DD252 pKa = 3.07IPKK255 pKa = 8.76ICKK258 pKa = 9.96VNFKK262 pKa = 10.86INFTEE267 pKa = 3.93
MM1 pKa = 7.33SWRR4 pKa = 11.84PPAYY8 pKa = 10.19NLAQRR13 pKa = 11.84EE14 pKa = 4.27EE15 pKa = 4.03HH16 pKa = 6.36WYY18 pKa = 10.05KK19 pKa = 11.02AILEE23 pKa = 4.29SHH25 pKa = 5.85STFCGCSDD33 pKa = 3.83VVRR36 pKa = 11.84HH37 pKa = 5.51FCVLASRR44 pKa = 11.84LATNPSVIPALPAPPEE60 pKa = 3.94QQPRR64 pKa = 11.84PGGDD68 pKa = 3.09TEE70 pKa = 4.61GAPGDD75 pKa = 4.46PGDD78 pKa = 4.48AGAGRR83 pKa = 11.84YY84 pKa = 9.19AEE86 pKa = 4.5EE87 pKa = 4.4DD88 pKa = 3.66LEE90 pKa = 4.37EE91 pKa = 4.54LFAAAAKK98 pKa = 10.66DD99 pKa = 3.58DD100 pKa = 3.62MRR102 pKa = 11.84RR103 pKa = 11.84VQKK106 pKa = 9.49TPNGNPVRR114 pKa = 11.84GSHH117 pKa = 6.1RR118 pKa = 11.84KK119 pKa = 7.58PRR121 pKa = 11.84RR122 pKa = 11.84RR123 pKa = 11.84RR124 pKa = 11.84LLFTQSTPKK133 pKa = 10.18RR134 pKa = 11.84ARR136 pKa = 11.84EE137 pKa = 3.75QPVRR141 pKa = 11.84RR142 pKa = 11.84RR143 pKa = 11.84DD144 pKa = 3.2SPRR147 pKa = 11.84RR148 pKa = 11.84GDD150 pKa = 3.53PKK152 pKa = 10.57TATTQAAQPAATAAASPQRR171 pKa = 11.84GTQTSPGRR179 pKa = 11.84RR180 pKa = 11.84PPTPEE185 pKa = 4.17RR186 pKa = 11.84SPLGPPPIIIQGDD199 pKa = 4.68PIPDD203 pKa = 3.78LLFPSTGKK211 pKa = 9.8KK212 pKa = 9.82RR213 pKa = 11.84KK214 pKa = 8.94FSKK217 pKa = 10.58FDD219 pKa = 3.19WEE221 pKa = 4.34TEE223 pKa = 3.97AEE225 pKa = 3.77IGRR228 pKa = 11.84WLRR231 pKa = 11.84RR232 pKa = 11.84PMRR235 pKa = 11.84FYY237 pKa = 10.66PSDD240 pKa = 3.84PPHH243 pKa = 6.43YY244 pKa = 9.4PWLPPKK250 pKa = 10.23RR251 pKa = 11.84DD252 pKa = 3.07IPKK255 pKa = 8.76ICKK258 pKa = 9.96VNFKK262 pKa = 10.86INFTEE267 pKa = 3.93
Molecular weight: 29.97 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1379 |
101 |
734 |
344.8 |
39.62 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.454 ± 1.981 | 1.668 ± 0.249 |
4.569 ± 0.446 | 5.511 ± 0.543 |
3.191 ± 0.536 | 5.656 ± 0.523 |
2.103 ± 0.116 | 3.481 ± 0.491 |
6.454 ± 0.758 | 7.034 ± 1.014 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.74 ± 0.426 | 4.206 ± 0.749 |
9.065 ± 1.942 | 4.714 ± 0.312 |
8.92 ± 1.147 | 7.977 ± 2.114 |
6.454 ± 0.479 | 3.626 ± 0.448 |
2.538 ± 0.409 | 4.641 ± 1.271 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
