
Simian foamy virus (isolate chimpanzee) (SFVcpz)
Taxonomy: Viruses; Riboviria; Pararnavirae; Artverviricota; Revtraviricetes; Ortervirales; Retroviridae; Spumaretrovirinae; Spumavirus; Simian foamy virus
Average proteome isoelectric point is 7.04
Get precalculated fractions of proteins
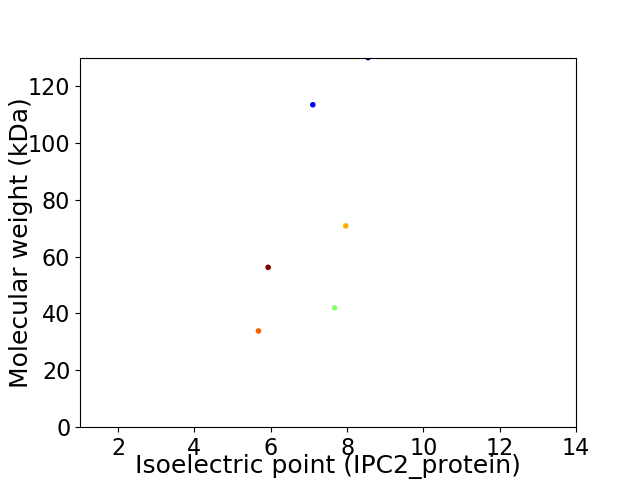
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
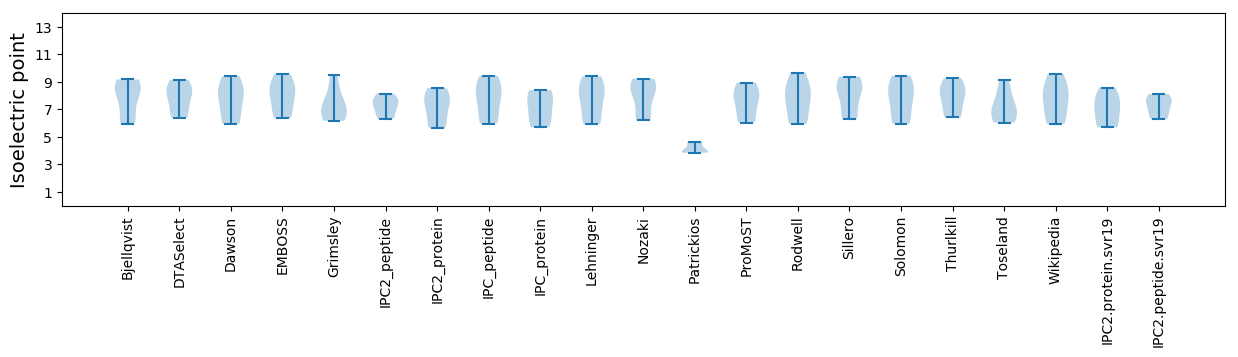
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q87043|BET_SFVCP Protein Bet OS=Simian foamy virus (isolate chimpanzee) OX=298339 GN=bet PE=3 SV=2
MM1 pKa = 8.15DD2 pKa = 5.26SYY4 pKa = 11.65QEE6 pKa = 4.13EE7 pKa = 4.57EE8 pKa = 4.45PVASTSGLQDD18 pKa = 3.54LQTLSEE24 pKa = 4.26LVGPEE29 pKa = 3.67NAGEE33 pKa = 4.19GDD35 pKa = 3.88LVIAEE40 pKa = 4.4EE41 pKa = 4.36PEE43 pKa = 4.05EE44 pKa = 4.12NPRR47 pKa = 11.84RR48 pKa = 11.84PRR50 pKa = 11.84RR51 pKa = 11.84YY52 pKa = 7.27TKK54 pKa = 10.46RR55 pKa = 11.84DD56 pKa = 3.58VKK58 pKa = 10.62CVSYY62 pKa = 10.26HH63 pKa = 6.49AYY65 pKa = 9.84KK66 pKa = 10.5EE67 pKa = 4.7LEE69 pKa = 4.62DD70 pKa = 3.87KK71 pKa = 10.9HH72 pKa = 5.92PHH74 pKa = 6.57HH75 pKa = 7.33IKK77 pKa = 10.4LQDD80 pKa = 3.88WIPKK84 pKa = 8.09PEE86 pKa = 4.05EE87 pKa = 3.78MSKK90 pKa = 10.64SICKK94 pKa = 10.12RR95 pKa = 11.84LILCGLYY102 pKa = 10.34SGEE105 pKa = 4.21KK106 pKa = 9.97ARR108 pKa = 11.84EE109 pKa = 3.79ILKK112 pKa = 10.73KK113 pKa = 10.03PFTVSWEE120 pKa = 4.03QSEE123 pKa = 4.6TNPDD127 pKa = 3.82CFIVSYY133 pKa = 9.84TCIFCDD139 pKa = 4.74AVIHH143 pKa = 7.18DD144 pKa = 4.8PMPVVWDD151 pKa = 3.82SEE153 pKa = 4.45VEE155 pKa = 3.58IWVKK159 pKa = 10.29YY160 pKa = 9.93KK161 pKa = 10.12PLRR164 pKa = 11.84GIVGSAVFIMEE175 pKa = 4.57KK176 pKa = 9.77HH177 pKa = 5.6QKK179 pKa = 9.38NCSLVKK185 pKa = 10.32PSTSCPEE192 pKa = 3.85GPKK195 pKa = 9.57PRR197 pKa = 11.84RR198 pKa = 11.84RR199 pKa = 11.84HH200 pKa = 6.23DD201 pKa = 3.31PVLRR205 pKa = 11.84CDD207 pKa = 3.51MFEE210 pKa = 3.97KK211 pKa = 10.12HH212 pKa = 6.29HH213 pKa = 6.51KK214 pKa = 9.38PRR216 pKa = 11.84PKK218 pKa = 9.97RR219 pKa = 11.84SRR221 pKa = 11.84KK222 pKa = 9.42RR223 pKa = 11.84SIDD226 pKa = 3.52HH227 pKa = 6.58EE228 pKa = 4.81SCASSGDD235 pKa = 3.68TVANEE240 pKa = 4.06SGPLCTNTFWTPGPVLQGLLGEE262 pKa = 4.63SSNLPDD268 pKa = 5.18LEE270 pKa = 4.3VHH272 pKa = 5.83MSGGPFWKK280 pKa = 9.88EE281 pKa = 3.62VYY283 pKa = 10.13GDD285 pKa = 4.47SILGPPSGSGEE296 pKa = 4.05HH297 pKa = 6.54SVLL300 pKa = 4.05
MM1 pKa = 8.15DD2 pKa = 5.26SYY4 pKa = 11.65QEE6 pKa = 4.13EE7 pKa = 4.57EE8 pKa = 4.45PVASTSGLQDD18 pKa = 3.54LQTLSEE24 pKa = 4.26LVGPEE29 pKa = 3.67NAGEE33 pKa = 4.19GDD35 pKa = 3.88LVIAEE40 pKa = 4.4EE41 pKa = 4.36PEE43 pKa = 4.05EE44 pKa = 4.12NPRR47 pKa = 11.84RR48 pKa = 11.84PRR50 pKa = 11.84RR51 pKa = 11.84YY52 pKa = 7.27TKK54 pKa = 10.46RR55 pKa = 11.84DD56 pKa = 3.58VKK58 pKa = 10.62CVSYY62 pKa = 10.26HH63 pKa = 6.49AYY65 pKa = 9.84KK66 pKa = 10.5EE67 pKa = 4.7LEE69 pKa = 4.62DD70 pKa = 3.87KK71 pKa = 10.9HH72 pKa = 5.92PHH74 pKa = 6.57HH75 pKa = 7.33IKK77 pKa = 10.4LQDD80 pKa = 3.88WIPKK84 pKa = 8.09PEE86 pKa = 4.05EE87 pKa = 3.78MSKK90 pKa = 10.64SICKK94 pKa = 10.12RR95 pKa = 11.84LILCGLYY102 pKa = 10.34SGEE105 pKa = 4.21KK106 pKa = 9.97ARR108 pKa = 11.84EE109 pKa = 3.79ILKK112 pKa = 10.73KK113 pKa = 10.03PFTVSWEE120 pKa = 4.03QSEE123 pKa = 4.6TNPDD127 pKa = 3.82CFIVSYY133 pKa = 9.84TCIFCDD139 pKa = 4.74AVIHH143 pKa = 7.18DD144 pKa = 4.8PMPVVWDD151 pKa = 3.82SEE153 pKa = 4.45VEE155 pKa = 3.58IWVKK159 pKa = 10.29YY160 pKa = 9.93KK161 pKa = 10.12PLRR164 pKa = 11.84GIVGSAVFIMEE175 pKa = 4.57KK176 pKa = 9.77HH177 pKa = 5.6QKK179 pKa = 9.38NCSLVKK185 pKa = 10.32PSTSCPEE192 pKa = 3.85GPKK195 pKa = 9.57PRR197 pKa = 11.84RR198 pKa = 11.84RR199 pKa = 11.84HH200 pKa = 6.23DD201 pKa = 3.31PVLRR205 pKa = 11.84CDD207 pKa = 3.51MFEE210 pKa = 3.97KK211 pKa = 10.12HH212 pKa = 6.29HH213 pKa = 6.51KK214 pKa = 9.38PRR216 pKa = 11.84PKK218 pKa = 9.97RR219 pKa = 11.84SRR221 pKa = 11.84KK222 pKa = 9.42RR223 pKa = 11.84SIDD226 pKa = 3.52HH227 pKa = 6.58EE228 pKa = 4.81SCASSGDD235 pKa = 3.68TVANEE240 pKa = 4.06SGPLCTNTFWTPGPVLQGLLGEE262 pKa = 4.63SSNLPDD268 pKa = 5.18LEE270 pKa = 4.3VHH272 pKa = 5.83MSGGPFWKK280 pKa = 9.88EE281 pKa = 3.62VYY283 pKa = 10.13GDD285 pKa = 4.47SILGPPSGSGEE296 pKa = 4.05HH297 pKa = 6.54SVLL300 pKa = 4.05
Molecular weight: 33.83 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q87041|ENV_SFVCP Envelope glycoprotein gp130 OS=Simian foamy virus (isolate chimpanzee) OX=298339 GN=env PE=1 SV=1
MM1 pKa = 7.58NPLQLLQPLPAEE13 pKa = 4.36VKK15 pKa = 8.81GTKK18 pKa = 10.06LLAHH22 pKa = 7.26WDD24 pKa = 3.38SGATITCIPEE34 pKa = 3.91SFLEE38 pKa = 4.26DD39 pKa = 3.61EE40 pKa = 4.51QPIKK44 pKa = 9.74QTLIKK49 pKa = 9.02TIHH52 pKa = 6.5GEE54 pKa = 3.94KK55 pKa = 9.11QQNVYY60 pKa = 11.11YY61 pKa = 9.72LTFKK65 pKa = 10.78VKK67 pKa = 9.58GRR69 pKa = 11.84KK70 pKa = 8.78VEE72 pKa = 4.14AEE74 pKa = 4.43VIASPYY80 pKa = 10.53EE81 pKa = 4.08YY82 pKa = 10.19ILLSPTDD89 pKa = 4.22VPWLTQQPLQLTILVPLQEE108 pKa = 4.3YY109 pKa = 9.4QDD111 pKa = 4.48RR112 pKa = 11.84ILNKK116 pKa = 8.67TALPEE121 pKa = 3.99EE122 pKa = 4.49QKK124 pKa = 10.35QQLKK128 pKa = 10.88ALFTKK133 pKa = 10.53YY134 pKa = 10.87DD135 pKa = 3.77NLWQHH140 pKa = 6.27WEE142 pKa = 3.98NQVGHH147 pKa = 6.44RR148 pKa = 11.84KK149 pKa = 8.88IRR151 pKa = 11.84PHH153 pKa = 6.76NIATGDD159 pKa = 3.71YY160 pKa = 9.61PPRR163 pKa = 11.84PQKK166 pKa = 10.35QYY168 pKa = 10.69PINPKK173 pKa = 9.94AKK175 pKa = 9.26PSIQIVIDD183 pKa = 4.01DD184 pKa = 4.33LLKK187 pKa = 10.97QGVLTPQNSTMNTPVYY203 pKa = 9.51PVPKK207 pKa = 9.66PDD209 pKa = 3.26GRR211 pKa = 11.84WRR213 pKa = 11.84MVLDD217 pKa = 3.37YY218 pKa = 11.29RR219 pKa = 11.84EE220 pKa = 4.13VNKK223 pKa = 9.63TIPLTAAQNQHH234 pKa = 5.59SAGILATIVRR244 pKa = 11.84QKK246 pKa = 11.38YY247 pKa = 7.57KK248 pKa = 8.69TTLDD252 pKa = 3.55LANGFWAHH260 pKa = 7.52PITPDD265 pKa = 4.31SYY267 pKa = 10.69WLTAFTWQGKK277 pKa = 6.31QYY279 pKa = 10.3CWTRR283 pKa = 11.84LPQGFLNSPALFTADD298 pKa = 4.28AVDD301 pKa = 4.11LLKK304 pKa = 10.55EE305 pKa = 4.11VPNVQVYY312 pKa = 10.52VDD314 pKa = 5.3DD315 pKa = 5.29IYY317 pKa = 11.41LSHH320 pKa = 7.73DD321 pKa = 3.69NPHH324 pKa = 5.64EE325 pKa = 4.83HH326 pKa = 6.19IQQLEE331 pKa = 4.22KK332 pKa = 10.97VFQILLQAGYY342 pKa = 10.34VVSLKK347 pKa = 10.48KK348 pKa = 10.57SEE350 pKa = 3.66IGQRR354 pKa = 11.84TVEE357 pKa = 3.87FLGFNITKK365 pKa = 9.84EE366 pKa = 4.23GRR368 pKa = 11.84GLTDD372 pKa = 3.13TFKK375 pKa = 10.78TKK377 pKa = 10.49LLNVTPPKK385 pKa = 10.22DD386 pKa = 3.68LKK388 pKa = 10.75QLQSILGLLNFARR401 pKa = 11.84NFIPNFAEE409 pKa = 4.98LVQTLYY415 pKa = 11.6NLIASSKK422 pKa = 9.77GKK424 pKa = 10.09YY425 pKa = 9.47IEE427 pKa = 4.01WTEE430 pKa = 4.81DD431 pKa = 2.9NTKK434 pKa = 10.32QLNKK438 pKa = 10.1VIEE441 pKa = 4.49ALNTASNLEE450 pKa = 3.94EE451 pKa = 4.21RR452 pKa = 11.84LPDD455 pKa = 3.22QRR457 pKa = 11.84LVIKK461 pKa = 11.0VNTSPSAGYY470 pKa = 9.01VRR472 pKa = 11.84YY473 pKa = 10.31YY474 pKa = 10.82NEE476 pKa = 4.28SGKK479 pKa = 10.57KK480 pKa = 9.46PIMYY484 pKa = 10.05LNYY487 pKa = 9.51VFSKK491 pKa = 11.39AEE493 pKa = 4.14LKK495 pKa = 10.77FSMLEE500 pKa = 3.85KK501 pKa = 10.79LLTTMHH507 pKa = 6.98KK508 pKa = 10.82ALIKK512 pKa = 11.07AMDD515 pKa = 3.95LAMGQEE521 pKa = 3.93ILVYY525 pKa = 10.46SPIVSMTKK533 pKa = 9.42IQKK536 pKa = 8.42TPLPEE541 pKa = 4.38RR542 pKa = 11.84KK543 pKa = 9.23ALPIRR548 pKa = 11.84WITWMTYY555 pKa = 10.7LEE557 pKa = 4.43DD558 pKa = 3.7PRR560 pKa = 11.84IQFHH564 pKa = 5.99YY565 pKa = 10.92DD566 pKa = 2.7KK567 pKa = 10.5TLPEE571 pKa = 4.67LKK573 pKa = 10.03HH574 pKa = 7.56IPDD577 pKa = 4.49VYY579 pKa = 10.31TSSIPPLKK587 pKa = 10.55HH588 pKa = 5.65PSQYY592 pKa = 11.05EE593 pKa = 4.21GVFCTDD599 pKa = 2.76GSAIKK604 pKa = 10.81SPDD607 pKa = 3.28PTKK610 pKa = 11.16SNNAGMGIVHH620 pKa = 7.8AIYY623 pKa = 10.46NPEE626 pKa = 3.9YY627 pKa = 10.58KK628 pKa = 9.94ILNQWSIPLGHH639 pKa = 6.84HH640 pKa = 5.71TAQMAEE646 pKa = 3.68IAAVEE651 pKa = 4.32FACKK655 pKa = 9.8KK656 pKa = 9.41ALKK659 pKa = 10.53VPGPVLVITDD669 pKa = 3.76SFYY672 pKa = 11.32VAEE675 pKa = 4.48SANKK679 pKa = 8.91EE680 pKa = 3.85LPYY683 pKa = 9.92WKK685 pKa = 10.68SNGFVNNKK693 pKa = 9.6KK694 pKa = 10.33EE695 pKa = 4.04PLKK698 pKa = 10.84HH699 pKa = 5.17ISKK702 pKa = 9.07WKK704 pKa = 10.54SIAEE708 pKa = 4.14CLSIKK713 pKa = 10.25PDD715 pKa = 2.96ITIQHH720 pKa = 6.25EE721 pKa = 4.58KK722 pKa = 9.8GHH724 pKa = 5.53QPINTSIHH732 pKa = 5.56TEE734 pKa = 4.09GNALADD740 pKa = 3.72KK741 pKa = 10.67LATQGSYY748 pKa = 9.31VVNCNTKK755 pKa = 10.23KK756 pKa = 10.43PNLDD760 pKa = 3.67AEE762 pKa = 4.72LDD764 pKa = 3.59QLLQGNNVKK773 pKa = 10.3GYY775 pKa = 8.0PKK777 pKa = 10.18QYY779 pKa = 10.25TYY781 pKa = 11.36YY782 pKa = 11.02LEE784 pKa = 4.6DD785 pKa = 3.48GKK787 pKa = 11.54VKK789 pKa = 10.2VSRR792 pKa = 11.84PEE794 pKa = 3.75GVKK797 pKa = 10.18IIPPQSDD804 pKa = 3.12RR805 pKa = 11.84QKK807 pKa = 10.81IVLQAHH813 pKa = 6.08NLAHH817 pKa = 6.87TGRR820 pKa = 11.84EE821 pKa = 3.94ATLLKK826 pKa = 10.2IANLYY831 pKa = 6.99WWPNMRR837 pKa = 11.84KK838 pKa = 9.79DD839 pKa = 3.63VVKK842 pKa = 10.75QLGRR846 pKa = 11.84CKK848 pKa = 10.27QCLITNASNKK858 pKa = 8.96TSGPILRR865 pKa = 11.84PDD867 pKa = 3.88RR868 pKa = 11.84PQKK871 pKa = 10.66PFDD874 pKa = 4.08KK875 pKa = 10.93FFIDD879 pKa = 4.29YY880 pKa = 10.24IGPLPPSQGYY890 pKa = 10.06LYY892 pKa = 11.05VLVIVDD898 pKa = 4.14GMTGFTWLYY907 pKa = 5.78PTKK910 pKa = 10.57APSTSATVKK919 pKa = 10.31SLNVLTSIAIPKK931 pKa = 9.28VIHH934 pKa = 6.65SDD936 pKa = 2.79QGAAFTSSTFAEE948 pKa = 4.41WAKK951 pKa = 10.58EE952 pKa = 3.85RR953 pKa = 11.84GIHH956 pKa = 6.76LEE958 pKa = 3.71FSTPYY963 pKa = 10.08HH964 pKa = 5.98PQSSGKK970 pKa = 8.3VEE972 pKa = 4.3RR973 pKa = 11.84KK974 pKa = 10.0NSDD977 pKa = 2.61IKK979 pKa = 11.08RR980 pKa = 11.84LLTKK984 pKa = 10.58LLVGRR989 pKa = 11.84PTKK992 pKa = 10.03WYY994 pKa = 9.91DD995 pKa = 3.56LLPVVQLALNNTYY1008 pKa = 10.92SPVLKK1013 pKa = 9.14YY1014 pKa = 9.21TPHH1017 pKa = 6.16QLLFGIDD1024 pKa = 3.66SNTPFANQDD1033 pKa = 3.49TLDD1036 pKa = 3.66LTRR1039 pKa = 11.84EE1040 pKa = 4.18EE1041 pKa = 4.48EE1042 pKa = 4.13LSLLQEE1048 pKa = 4.32IRR1050 pKa = 11.84ASLYY1054 pKa = 10.43QPSTPPASSRR1064 pKa = 11.84SWSPVVGQLVQEE1076 pKa = 4.66RR1077 pKa = 11.84VARR1080 pKa = 11.84PASLRR1085 pKa = 11.84PRR1087 pKa = 11.84WHH1089 pKa = 6.64KK1090 pKa = 10.31PSTVLEE1096 pKa = 4.18VLNPRR1101 pKa = 11.84TVVILDD1107 pKa = 3.34HH1108 pKa = 6.8LGNNRR1113 pKa = 11.84TVSIDD1118 pKa = 3.34NLKK1121 pKa = 8.75PTSHH1125 pKa = 6.65QNGTTNDD1132 pKa = 3.86TATMDD1137 pKa = 3.53HH1138 pKa = 6.93LEE1140 pKa = 4.06QNEE1143 pKa = 4.06QSSS1146 pKa = 3.61
MM1 pKa = 7.58NPLQLLQPLPAEE13 pKa = 4.36VKK15 pKa = 8.81GTKK18 pKa = 10.06LLAHH22 pKa = 7.26WDD24 pKa = 3.38SGATITCIPEE34 pKa = 3.91SFLEE38 pKa = 4.26DD39 pKa = 3.61EE40 pKa = 4.51QPIKK44 pKa = 9.74QTLIKK49 pKa = 9.02TIHH52 pKa = 6.5GEE54 pKa = 3.94KK55 pKa = 9.11QQNVYY60 pKa = 11.11YY61 pKa = 9.72LTFKK65 pKa = 10.78VKK67 pKa = 9.58GRR69 pKa = 11.84KK70 pKa = 8.78VEE72 pKa = 4.14AEE74 pKa = 4.43VIASPYY80 pKa = 10.53EE81 pKa = 4.08YY82 pKa = 10.19ILLSPTDD89 pKa = 4.22VPWLTQQPLQLTILVPLQEE108 pKa = 4.3YY109 pKa = 9.4QDD111 pKa = 4.48RR112 pKa = 11.84ILNKK116 pKa = 8.67TALPEE121 pKa = 3.99EE122 pKa = 4.49QKK124 pKa = 10.35QQLKK128 pKa = 10.88ALFTKK133 pKa = 10.53YY134 pKa = 10.87DD135 pKa = 3.77NLWQHH140 pKa = 6.27WEE142 pKa = 3.98NQVGHH147 pKa = 6.44RR148 pKa = 11.84KK149 pKa = 8.88IRR151 pKa = 11.84PHH153 pKa = 6.76NIATGDD159 pKa = 3.71YY160 pKa = 9.61PPRR163 pKa = 11.84PQKK166 pKa = 10.35QYY168 pKa = 10.69PINPKK173 pKa = 9.94AKK175 pKa = 9.26PSIQIVIDD183 pKa = 4.01DD184 pKa = 4.33LLKK187 pKa = 10.97QGVLTPQNSTMNTPVYY203 pKa = 9.51PVPKK207 pKa = 9.66PDD209 pKa = 3.26GRR211 pKa = 11.84WRR213 pKa = 11.84MVLDD217 pKa = 3.37YY218 pKa = 11.29RR219 pKa = 11.84EE220 pKa = 4.13VNKK223 pKa = 9.63TIPLTAAQNQHH234 pKa = 5.59SAGILATIVRR244 pKa = 11.84QKK246 pKa = 11.38YY247 pKa = 7.57KK248 pKa = 8.69TTLDD252 pKa = 3.55LANGFWAHH260 pKa = 7.52PITPDD265 pKa = 4.31SYY267 pKa = 10.69WLTAFTWQGKK277 pKa = 6.31QYY279 pKa = 10.3CWTRR283 pKa = 11.84LPQGFLNSPALFTADD298 pKa = 4.28AVDD301 pKa = 4.11LLKK304 pKa = 10.55EE305 pKa = 4.11VPNVQVYY312 pKa = 10.52VDD314 pKa = 5.3DD315 pKa = 5.29IYY317 pKa = 11.41LSHH320 pKa = 7.73DD321 pKa = 3.69NPHH324 pKa = 5.64EE325 pKa = 4.83HH326 pKa = 6.19IQQLEE331 pKa = 4.22KK332 pKa = 10.97VFQILLQAGYY342 pKa = 10.34VVSLKK347 pKa = 10.48KK348 pKa = 10.57SEE350 pKa = 3.66IGQRR354 pKa = 11.84TVEE357 pKa = 3.87FLGFNITKK365 pKa = 9.84EE366 pKa = 4.23GRR368 pKa = 11.84GLTDD372 pKa = 3.13TFKK375 pKa = 10.78TKK377 pKa = 10.49LLNVTPPKK385 pKa = 10.22DD386 pKa = 3.68LKK388 pKa = 10.75QLQSILGLLNFARR401 pKa = 11.84NFIPNFAEE409 pKa = 4.98LVQTLYY415 pKa = 11.6NLIASSKK422 pKa = 9.77GKK424 pKa = 10.09YY425 pKa = 9.47IEE427 pKa = 4.01WTEE430 pKa = 4.81DD431 pKa = 2.9NTKK434 pKa = 10.32QLNKK438 pKa = 10.1VIEE441 pKa = 4.49ALNTASNLEE450 pKa = 3.94EE451 pKa = 4.21RR452 pKa = 11.84LPDD455 pKa = 3.22QRR457 pKa = 11.84LVIKK461 pKa = 11.0VNTSPSAGYY470 pKa = 9.01VRR472 pKa = 11.84YY473 pKa = 10.31YY474 pKa = 10.82NEE476 pKa = 4.28SGKK479 pKa = 10.57KK480 pKa = 9.46PIMYY484 pKa = 10.05LNYY487 pKa = 9.51VFSKK491 pKa = 11.39AEE493 pKa = 4.14LKK495 pKa = 10.77FSMLEE500 pKa = 3.85KK501 pKa = 10.79LLTTMHH507 pKa = 6.98KK508 pKa = 10.82ALIKK512 pKa = 11.07AMDD515 pKa = 3.95LAMGQEE521 pKa = 3.93ILVYY525 pKa = 10.46SPIVSMTKK533 pKa = 9.42IQKK536 pKa = 8.42TPLPEE541 pKa = 4.38RR542 pKa = 11.84KK543 pKa = 9.23ALPIRR548 pKa = 11.84WITWMTYY555 pKa = 10.7LEE557 pKa = 4.43DD558 pKa = 3.7PRR560 pKa = 11.84IQFHH564 pKa = 5.99YY565 pKa = 10.92DD566 pKa = 2.7KK567 pKa = 10.5TLPEE571 pKa = 4.67LKK573 pKa = 10.03HH574 pKa = 7.56IPDD577 pKa = 4.49VYY579 pKa = 10.31TSSIPPLKK587 pKa = 10.55HH588 pKa = 5.65PSQYY592 pKa = 11.05EE593 pKa = 4.21GVFCTDD599 pKa = 2.76GSAIKK604 pKa = 10.81SPDD607 pKa = 3.28PTKK610 pKa = 11.16SNNAGMGIVHH620 pKa = 7.8AIYY623 pKa = 10.46NPEE626 pKa = 3.9YY627 pKa = 10.58KK628 pKa = 9.94ILNQWSIPLGHH639 pKa = 6.84HH640 pKa = 5.71TAQMAEE646 pKa = 3.68IAAVEE651 pKa = 4.32FACKK655 pKa = 9.8KK656 pKa = 9.41ALKK659 pKa = 10.53VPGPVLVITDD669 pKa = 3.76SFYY672 pKa = 11.32VAEE675 pKa = 4.48SANKK679 pKa = 8.91EE680 pKa = 3.85LPYY683 pKa = 9.92WKK685 pKa = 10.68SNGFVNNKK693 pKa = 9.6KK694 pKa = 10.33EE695 pKa = 4.04PLKK698 pKa = 10.84HH699 pKa = 5.17ISKK702 pKa = 9.07WKK704 pKa = 10.54SIAEE708 pKa = 4.14CLSIKK713 pKa = 10.25PDD715 pKa = 2.96ITIQHH720 pKa = 6.25EE721 pKa = 4.58KK722 pKa = 9.8GHH724 pKa = 5.53QPINTSIHH732 pKa = 5.56TEE734 pKa = 4.09GNALADD740 pKa = 3.72KK741 pKa = 10.67LATQGSYY748 pKa = 9.31VVNCNTKK755 pKa = 10.23KK756 pKa = 10.43PNLDD760 pKa = 3.67AEE762 pKa = 4.72LDD764 pKa = 3.59QLLQGNNVKK773 pKa = 10.3GYY775 pKa = 8.0PKK777 pKa = 10.18QYY779 pKa = 10.25TYY781 pKa = 11.36YY782 pKa = 11.02LEE784 pKa = 4.6DD785 pKa = 3.48GKK787 pKa = 11.54VKK789 pKa = 10.2VSRR792 pKa = 11.84PEE794 pKa = 3.75GVKK797 pKa = 10.18IIPPQSDD804 pKa = 3.12RR805 pKa = 11.84QKK807 pKa = 10.81IVLQAHH813 pKa = 6.08NLAHH817 pKa = 6.87TGRR820 pKa = 11.84EE821 pKa = 3.94ATLLKK826 pKa = 10.2IANLYY831 pKa = 6.99WWPNMRR837 pKa = 11.84KK838 pKa = 9.79DD839 pKa = 3.63VVKK842 pKa = 10.75QLGRR846 pKa = 11.84CKK848 pKa = 10.27QCLITNASNKK858 pKa = 8.96TSGPILRR865 pKa = 11.84PDD867 pKa = 3.88RR868 pKa = 11.84PQKK871 pKa = 10.66PFDD874 pKa = 4.08KK875 pKa = 10.93FFIDD879 pKa = 4.29YY880 pKa = 10.24IGPLPPSQGYY890 pKa = 10.06LYY892 pKa = 11.05VLVIVDD898 pKa = 4.14GMTGFTWLYY907 pKa = 5.78PTKK910 pKa = 10.57APSTSATVKK919 pKa = 10.31SLNVLTSIAIPKK931 pKa = 9.28VIHH934 pKa = 6.65SDD936 pKa = 2.79QGAAFTSSTFAEE948 pKa = 4.41WAKK951 pKa = 10.58EE952 pKa = 3.85RR953 pKa = 11.84GIHH956 pKa = 6.76LEE958 pKa = 3.71FSTPYY963 pKa = 10.08HH964 pKa = 5.98PQSSGKK970 pKa = 8.3VEE972 pKa = 4.3RR973 pKa = 11.84KK974 pKa = 10.0NSDD977 pKa = 2.61IKK979 pKa = 11.08RR980 pKa = 11.84LLTKK984 pKa = 10.58LLVGRR989 pKa = 11.84PTKK992 pKa = 10.03WYY994 pKa = 9.91DD995 pKa = 3.56LLPVVQLALNNTYY1008 pKa = 10.92SPVLKK1013 pKa = 9.14YY1014 pKa = 9.21TPHH1017 pKa = 6.16QLLFGIDD1024 pKa = 3.66SNTPFANQDD1033 pKa = 3.49TLDD1036 pKa = 3.66LTRR1039 pKa = 11.84EE1040 pKa = 4.18EE1041 pKa = 4.48EE1042 pKa = 4.13LSLLQEE1048 pKa = 4.32IRR1050 pKa = 11.84ASLYY1054 pKa = 10.43QPSTPPASSRR1064 pKa = 11.84SWSPVVGQLVQEE1076 pKa = 4.66RR1077 pKa = 11.84VARR1080 pKa = 11.84PASLRR1085 pKa = 11.84PRR1087 pKa = 11.84WHH1089 pKa = 6.64KK1090 pKa = 10.31PSTVLEE1096 pKa = 4.18VLNPRR1101 pKa = 11.84TVVILDD1107 pKa = 3.34HH1108 pKa = 6.8LGNNRR1113 pKa = 11.84TVSIDD1118 pKa = 3.34NLKK1121 pKa = 8.75PTSHH1125 pKa = 6.65QNGTTNDD1132 pKa = 3.86TATMDD1137 pKa = 3.53HH1138 pKa = 6.93LEE1140 pKa = 4.06QNEE1143 pKa = 4.06QSSS1146 pKa = 3.61
Molecular weight: 129.93 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
3941 |
300 |
1146 |
656.8 |
74.35 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.633 ± 0.341 | 1.675 ± 0.502 |
4.517 ± 0.384 | 6.09 ± 0.473 |
2.512 ± 0.183 | 6.344 ± 1.077 |
2.639 ± 0.196 | 5.684 ± 0.57 |
6.064 ± 1.077 | 8.932 ± 0.829 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.827 ± 0.188 | 4.796 ± 0.385 |
7.435 ± 0.764 | 5.785 ± 0.403 |
5.151 ± 0.899 | 6.623 ± 0.587 |
5.963 ± 0.456 | 5.938 ± 0.463 |
2.005 ± 0.295 | 4.39 ± 0.539 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
