
Janibacter sp. HTCC2649
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Intrasporangiaceae; Janibacter; unclassified Janibacter
Average proteome isoelectric point is 6.09
Get precalculated fractions of proteins
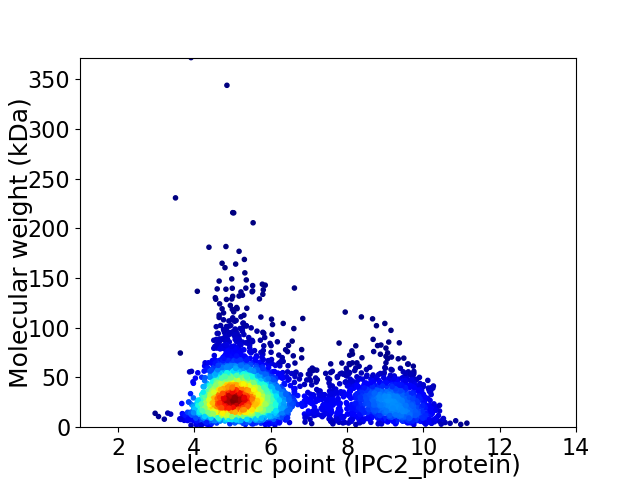
Virtual 2D-PAGE plot for 4081 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
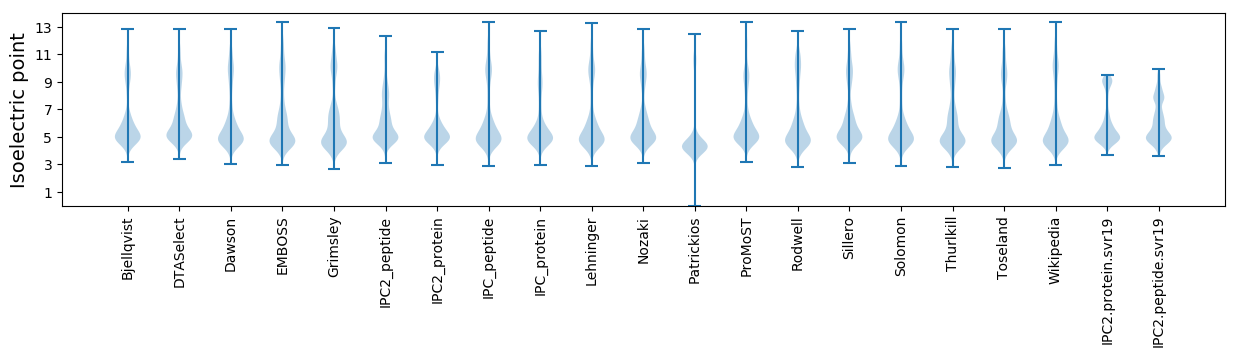
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A3TLQ1|A3TLQ1_9MICO Carbamoyl-phosphate synthase small chain OS=Janibacter sp. HTCC2649 OX=313589 GN=carA PE=3 SV=1
MM1 pKa = 7.75LSRR4 pKa = 11.84WCALTGIGVVVAAMGLTPSAAAAPYY29 pKa = 10.0AAVASVSPGVQYY41 pKa = 11.09LGDD44 pKa = 3.8STGVTFTFMVANTGTADD61 pKa = 3.43SIGAVEE67 pKa = 4.33IDD69 pKa = 3.53RR70 pKa = 11.84PGKK73 pKa = 8.52QWQITGCPSAPTGWSAQRR91 pKa = 11.84SDD93 pKa = 3.32TACRR97 pKa = 11.84FRR99 pKa = 11.84SAAGTGDD106 pKa = 4.79DD107 pKa = 4.45IAPGQQTTAFRR118 pKa = 11.84VTAVVAPGTQNLTGTWPVKK137 pKa = 10.43VSRR140 pKa = 11.84SSNFDD145 pKa = 3.34TKK147 pKa = 11.25SKK149 pKa = 10.71LIAAASTSPGLGITAHH165 pKa = 6.48SFQILDD171 pKa = 3.69AVVDD175 pKa = 4.08PATSTVGSACPSPAKK190 pKa = 10.45SAVTGSAGHH199 pKa = 6.56TIVICGRR206 pKa = 11.84NRR208 pKa = 11.84TTGTLTPTAAQSSLGGTFLAGQGAFSSGPVAPTTASVVLGRR249 pKa = 11.84WSNVTITTLAGPGKK263 pKa = 8.44TVIAKK268 pKa = 10.0IGSATNRR275 pKa = 11.84TSPLTTLTGYY285 pKa = 7.46TALNTPPNASDD296 pKa = 3.63DD297 pKa = 3.78AVTIDD302 pKa = 4.39EE303 pKa = 5.0DD304 pKa = 4.33SSTSFDD310 pKa = 3.55PRR312 pKa = 11.84TNDD315 pKa = 3.52SDD317 pKa = 4.31PDD319 pKa = 3.55GDD321 pKa = 4.39AFTISAIGTTGTQGAVTITGGGTGLTYY348 pKa = 10.95DD349 pKa = 4.34PDD351 pKa = 3.98GKK353 pKa = 10.6FDD355 pKa = 3.7NLAPGEE361 pKa = 4.08QDD363 pKa = 2.6TDD365 pKa = 3.5TFTYY369 pKa = 9.96TITDD373 pKa = 3.33AFGGSDD379 pKa = 3.34SATVTVTVTGLADD392 pKa = 3.56PPVAVNDD399 pKa = 3.87SKK401 pKa = 10.21TVVEE405 pKa = 4.84DD406 pKa = 4.29APATTVDD413 pKa = 3.53VLANDD418 pKa = 3.87TDD420 pKa = 3.88VDD422 pKa = 4.1GGPMSIASVTQPANGTVVITGGGTGLTYY450 pKa = 10.64EE451 pKa = 4.81PDD453 pKa = 3.14AGYY456 pKa = 10.39CNSVAGPADD465 pKa = 3.73PFTYY469 pKa = 10.04TLAPGGSSATVSVTVTCVDD488 pKa = 4.21DD489 pKa = 5.03LPVAVDD495 pKa = 3.55DD496 pKa = 4.7AKK498 pKa = 10.56TVVEE502 pKa = 4.71DD503 pKa = 3.93AAATAVDD510 pKa = 3.93VLANDD515 pKa = 3.87TDD517 pKa = 3.85VDD519 pKa = 3.88GGPISIASVTQPANGTVVITGGGTGLTYY547 pKa = 10.67QPDD550 pKa = 3.23ASYY553 pKa = 11.5CNNPPGTTPDD563 pKa = 3.27TFTYY567 pKa = 9.99TLTPGGSTAIVSVTVTCVDD586 pKa = 4.21DD587 pKa = 5.03LPVAVDD593 pKa = 3.59DD594 pKa = 5.01AKK596 pKa = 10.7TLVEE600 pKa = 4.54DD601 pKa = 4.36AAATAVDD608 pKa = 3.85VLVNDD613 pKa = 4.04TDD615 pKa = 3.66VDD617 pKa = 3.79GGPISITSVTQPTNGTVVITGGGTGLTYY645 pKa = 10.68QPDD648 pKa = 3.6ANYY651 pKa = 10.66CNNPPGTTLDD661 pKa = 3.42TFTYY665 pKa = 8.71TLSPGGSTATVSLTVTCVNDD685 pKa = 3.61PPNAVDD691 pKa = 3.58DD692 pKa = 4.58AKK694 pKa = 10.54TVVEE698 pKa = 4.71DD699 pKa = 3.93AAATAVDD706 pKa = 3.93VLANDD711 pKa = 4.13TDD713 pKa = 4.37PDD715 pKa = 4.13GDD717 pKa = 4.6PISITSVTQPANGTVAITGGGTGLTYY743 pKa = 10.71DD744 pKa = 4.41PNANYY749 pKa = 10.63CNDD752 pKa = 4.12PPGSTPDD759 pKa = 3.22TFTYY763 pKa = 9.12TVNGGSTGTVSVMVTCVDD781 pKa = 4.17DD782 pKa = 5.0LPVAVDD788 pKa = 4.07DD789 pKa = 4.85SEE791 pKa = 4.83QVTLNATATALMVLANDD808 pKa = 3.74TDD810 pKa = 3.73VDD812 pKa = 4.01GGALSIASVTQPTNGTVVITGGGTGLTYY840 pKa = 10.64EE841 pKa = 4.82PDD843 pKa = 3.11AGYY846 pKa = 10.96CDD848 pKa = 5.36DD849 pKa = 5.29PPGTTPDD856 pKa = 3.27TFTYY860 pKa = 9.17TLNGGSTAMVSVRR873 pKa = 11.84VVCDD877 pKa = 4.05DD878 pKa = 4.06PPVAVDD884 pKa = 3.52DD885 pKa = 4.57AKK887 pKa = 10.56TVVEE891 pKa = 4.71DD892 pKa = 3.93AAATAVDD899 pKa = 3.93VLANDD904 pKa = 3.87TDD906 pKa = 3.85VDD908 pKa = 3.88GGPISIASATQPTNGTVVITGGGTGLTYY936 pKa = 10.67QPDD939 pKa = 3.23ASYY942 pKa = 11.5CNNPPGTTLDD952 pKa = 3.47TFTYY956 pKa = 9.33TLTPGGSTATVSVTVTCVDD975 pKa = 3.83NLPVAVDD982 pKa = 3.58DD983 pKa = 4.63AKK985 pKa = 10.56TVVEE989 pKa = 4.71DD990 pKa = 3.93AAATAVDD997 pKa = 3.93VLANDD1002 pKa = 3.87TDD1004 pKa = 3.85VDD1006 pKa = 3.88GGPISIASVNQPTNGTVVISGGGTGLTYY1034 pKa = 10.68QPDD1037 pKa = 3.6ANYY1040 pKa = 10.66CNNPPGTTLDD1050 pKa = 3.47TFTYY1054 pKa = 9.33TLTPGGSTATVSLTVTCVNDD1074 pKa = 3.61PPNAVDD1080 pKa = 4.07DD1081 pKa = 4.09ATSVGEE1087 pKa = 4.36GAAGAAVPVLANDD1100 pKa = 3.94TDD1102 pKa = 4.45PDD1104 pKa = 3.95PTDD1107 pKa = 3.43VLSVTSVDD1115 pKa = 3.39TTGTIGGVSLVGGVVTYY1132 pKa = 10.87NPNGQFEE1139 pKa = 4.48YY1140 pKa = 10.65LAAGATATDD1149 pKa = 3.72TFSYY1153 pKa = 9.86TVSDD1157 pKa = 4.24GNGGTDD1163 pKa = 3.28TAVVTVTVTGANDD1176 pKa = 3.62PLVAVNDD1183 pKa = 3.94TGTTDD1188 pKa = 3.3EE1189 pKa = 5.06DD1190 pKa = 4.14TTLSQSAPGVLANDD1204 pKa = 4.02TDD1206 pKa = 4.75PDD1208 pKa = 3.85TGDD1211 pKa = 3.07TRR1213 pKa = 11.84TVTRR1217 pKa = 11.84LNGSATLTGTSTSGAAVSIAANGSFTYY1244 pKa = 10.7NPGGLFQGLSTGQSATDD1261 pKa = 3.41TFTYY1265 pKa = 9.08TAQDD1269 pKa = 3.29GSGAQSTATVTITINGVSDD1288 pKa = 3.76APTAVTDD1295 pKa = 3.81SFQTHH1300 pKa = 6.31GNTNLFVGTSRR1311 pKa = 11.84PVGEE1315 pKa = 4.4AGKK1318 pKa = 10.44VLTGSVLTNDD1328 pKa = 4.13TDD1330 pKa = 4.56PDD1332 pKa = 3.86TAQANLVAEE1341 pKa = 4.58PVTNAPTTLGGSITIEE1357 pKa = 3.61SDD1359 pKa = 3.62GNFTFYY1365 pKa = 10.47PDD1367 pKa = 4.81DD1368 pKa = 4.9GDD1370 pKa = 4.49SGVTDD1375 pKa = 3.41SFTYY1379 pKa = 9.9RR1380 pKa = 11.84VCDD1383 pKa = 4.02ASPCNASTVANATGTLNLPISSSQVWYY1410 pKa = 10.49VRR1412 pKa = 11.84NNAVAGGDD1420 pKa = 3.9GTSEE1424 pKa = 4.46APFDD1428 pKa = 3.93TLGEE1432 pKa = 4.27AEE1434 pKa = 4.25TAADD1438 pKa = 4.6AGDD1441 pKa = 3.52TTYY1444 pKa = 11.17VFDD1447 pKa = 5.75GDD1449 pKa = 3.88NTSLNLGSGFVMAAGEE1465 pKa = 4.19RR1466 pKa = 11.84LLGEE1470 pKa = 4.26AVTLQVGADD1479 pKa = 3.47VLYY1482 pKa = 10.47TGVPANRR1489 pKa = 11.84PTLTANDD1496 pKa = 3.84EE1497 pKa = 4.45DD1498 pKa = 4.9VVTLANATTVRR1509 pKa = 11.84GVQLDD1514 pKa = 3.73PQGAGGGIAGIGTASGPGTSTIADD1538 pKa = 3.29VRR1540 pKa = 11.84IIDD1543 pKa = 3.72TGTAGTQPGLEE1554 pKa = 4.05LDD1556 pKa = 4.17GVEE1559 pKa = 4.12GTYY1562 pKa = 10.34DD1563 pKa = 3.38ISDD1566 pKa = 3.73LTVDD1570 pKa = 3.73NSAATGQTAGSIGVRR1585 pKa = 11.84LNRR1588 pKa = 11.84NILGFTANFASAGTISLTTKK1608 pKa = 9.6GAKK1611 pKa = 9.82ALEE1614 pKa = 4.11VTGANMGLASVFDD1627 pKa = 5.71DD1628 pKa = 3.29ITVTGSPTGGVSMVNSLGSGTAFGDD1653 pKa = 4.11GVGVDD1658 pKa = 4.18LALTTTGGSAFNLQGTGTVSVPAAGTANVNATAGAGVFVNGSSGSSLAFDD1708 pKa = 4.26DD1709 pKa = 4.0VTVVSNPSGVGIAAVALGTGSFSAATGSISNTAGTAFTTGTGNGPISYY1757 pKa = 10.36GGTINNGSGRR1767 pKa = 11.84SVEE1770 pKa = 4.13VSARR1774 pKa = 11.84TGGTVTLTGNINDD1787 pKa = 4.15TNDD1790 pKa = 2.99VGGGIFVTGNSGGSTVLSGTTKK1812 pKa = 10.05TLNTGSEE1819 pKa = 4.2KK1820 pKa = 10.85AVDD1823 pKa = 4.58FSSSDD1828 pKa = 3.06GHH1830 pKa = 6.03TLTLSGGGLDD1840 pKa = 3.52IDD1842 pKa = 4.38TVSGTGVDD1850 pKa = 3.59AASSGTLNITGAGNTIDD1867 pKa = 3.64TSAGKK1872 pKa = 10.02ALSVSDD1878 pKa = 3.89TDD1880 pKa = 3.88VGPSPLSLQRR1890 pKa = 11.84ISSLGGAGITLSNTGSNNALVVASSGSGTCTAADD1924 pKa = 3.35QSGCTGGEE1932 pKa = 3.62IRR1934 pKa = 11.84SATGVDD1940 pKa = 3.81DD1941 pKa = 5.41SGALPTGTGIVLNNTRR1957 pKa = 11.84GVSLTRR1963 pKa = 11.84MHH1965 pKa = 6.8LHH1967 pKa = 6.38DD1968 pKa = 4.6HH1969 pKa = 6.11SNYY1972 pKa = 9.72GIRR1975 pKa = 11.84GTSVVGFSMVDD1986 pKa = 3.26SVVNGVNGTNGTTTFKK2002 pKa = 10.73DD2003 pKa = 3.62GSVRR2007 pKa = 11.84FVEE2010 pKa = 4.34LTGTVAMTNVALSGGYY2026 pKa = 7.55FTNLMVDD2033 pKa = 3.55NTAGILNGTFDD2044 pKa = 4.19NVDD2047 pKa = 3.37SGTINATGGDD2057 pKa = 3.66DD2058 pKa = 3.5AVQFEE2063 pKa = 5.29GIGTSTMNVDD2073 pKa = 4.1YY2074 pKa = 10.45KK2075 pKa = 11.32NSAIGTATGDD2085 pKa = 3.54LFQYY2089 pKa = 10.42IGDD2092 pKa = 3.94GTGGGNLDD2100 pKa = 3.54LTSNAFTNNEE2110 pKa = 3.75PSIATGGGGVAIVAGAKK2127 pKa = 9.64GAATLDD2133 pKa = 3.29IVSNTFRR2140 pKa = 11.84DD2141 pKa = 3.76SLTNALTVIKK2151 pKa = 10.57SADD2154 pKa = 3.3ATAGTNPLVSNITGNTIGVAATANSGSFEE2183 pKa = 4.23GDD2185 pKa = 2.72GMEE2188 pKa = 3.91ITTFGDD2194 pKa = 3.54GNATFNVLNNNISQYY2209 pKa = 10.39NSSGIQFVAGSGVTATGQMNLNVSGNVVGNPGTNPLITLLQGIRR2253 pKa = 11.84VDD2255 pKa = 4.04SGVDD2259 pKa = 3.15TADD2262 pKa = 3.41TFSTCVKK2269 pKa = 10.2FGANSISGAGGSAGRR2284 pKa = 11.84DD2285 pKa = 3.59FRR2287 pKa = 11.84LVASQQTTLRR2297 pKa = 11.84QPGYY2301 pKa = 10.82VGGSTDD2307 pKa = 2.99GTAFANYY2314 pKa = 8.74AASLIGSGAAGTAVANAPGTFSGAGSTCPP2343 pKa = 3.86
MM1 pKa = 7.75LSRR4 pKa = 11.84WCALTGIGVVVAAMGLTPSAAAAPYY29 pKa = 10.0AAVASVSPGVQYY41 pKa = 11.09LGDD44 pKa = 3.8STGVTFTFMVANTGTADD61 pKa = 3.43SIGAVEE67 pKa = 4.33IDD69 pKa = 3.53RR70 pKa = 11.84PGKK73 pKa = 8.52QWQITGCPSAPTGWSAQRR91 pKa = 11.84SDD93 pKa = 3.32TACRR97 pKa = 11.84FRR99 pKa = 11.84SAAGTGDD106 pKa = 4.79DD107 pKa = 4.45IAPGQQTTAFRR118 pKa = 11.84VTAVVAPGTQNLTGTWPVKK137 pKa = 10.43VSRR140 pKa = 11.84SSNFDD145 pKa = 3.34TKK147 pKa = 11.25SKK149 pKa = 10.71LIAAASTSPGLGITAHH165 pKa = 6.48SFQILDD171 pKa = 3.69AVVDD175 pKa = 4.08PATSTVGSACPSPAKK190 pKa = 10.45SAVTGSAGHH199 pKa = 6.56TIVICGRR206 pKa = 11.84NRR208 pKa = 11.84TTGTLTPTAAQSSLGGTFLAGQGAFSSGPVAPTTASVVLGRR249 pKa = 11.84WSNVTITTLAGPGKK263 pKa = 8.44TVIAKK268 pKa = 10.0IGSATNRR275 pKa = 11.84TSPLTTLTGYY285 pKa = 7.46TALNTPPNASDD296 pKa = 3.63DD297 pKa = 3.78AVTIDD302 pKa = 4.39EE303 pKa = 5.0DD304 pKa = 4.33SSTSFDD310 pKa = 3.55PRR312 pKa = 11.84TNDD315 pKa = 3.52SDD317 pKa = 4.31PDD319 pKa = 3.55GDD321 pKa = 4.39AFTISAIGTTGTQGAVTITGGGTGLTYY348 pKa = 10.95DD349 pKa = 4.34PDD351 pKa = 3.98GKK353 pKa = 10.6FDD355 pKa = 3.7NLAPGEE361 pKa = 4.08QDD363 pKa = 2.6TDD365 pKa = 3.5TFTYY369 pKa = 9.96TITDD373 pKa = 3.33AFGGSDD379 pKa = 3.34SATVTVTVTGLADD392 pKa = 3.56PPVAVNDD399 pKa = 3.87SKK401 pKa = 10.21TVVEE405 pKa = 4.84DD406 pKa = 4.29APATTVDD413 pKa = 3.53VLANDD418 pKa = 3.87TDD420 pKa = 3.88VDD422 pKa = 4.1GGPMSIASVTQPANGTVVITGGGTGLTYY450 pKa = 10.64EE451 pKa = 4.81PDD453 pKa = 3.14AGYY456 pKa = 10.39CNSVAGPADD465 pKa = 3.73PFTYY469 pKa = 10.04TLAPGGSSATVSVTVTCVDD488 pKa = 4.21DD489 pKa = 5.03LPVAVDD495 pKa = 3.55DD496 pKa = 4.7AKK498 pKa = 10.56TVVEE502 pKa = 4.71DD503 pKa = 3.93AAATAVDD510 pKa = 3.93VLANDD515 pKa = 3.87TDD517 pKa = 3.85VDD519 pKa = 3.88GGPISIASVTQPANGTVVITGGGTGLTYY547 pKa = 10.67QPDD550 pKa = 3.23ASYY553 pKa = 11.5CNNPPGTTPDD563 pKa = 3.27TFTYY567 pKa = 9.99TLTPGGSTAIVSVTVTCVDD586 pKa = 4.21DD587 pKa = 5.03LPVAVDD593 pKa = 3.59DD594 pKa = 5.01AKK596 pKa = 10.7TLVEE600 pKa = 4.54DD601 pKa = 4.36AAATAVDD608 pKa = 3.85VLVNDD613 pKa = 4.04TDD615 pKa = 3.66VDD617 pKa = 3.79GGPISITSVTQPTNGTVVITGGGTGLTYY645 pKa = 10.68QPDD648 pKa = 3.6ANYY651 pKa = 10.66CNNPPGTTLDD661 pKa = 3.42TFTYY665 pKa = 8.71TLSPGGSTATVSLTVTCVNDD685 pKa = 3.61PPNAVDD691 pKa = 3.58DD692 pKa = 4.58AKK694 pKa = 10.54TVVEE698 pKa = 4.71DD699 pKa = 3.93AAATAVDD706 pKa = 3.93VLANDD711 pKa = 4.13TDD713 pKa = 4.37PDD715 pKa = 4.13GDD717 pKa = 4.6PISITSVTQPANGTVAITGGGTGLTYY743 pKa = 10.71DD744 pKa = 4.41PNANYY749 pKa = 10.63CNDD752 pKa = 4.12PPGSTPDD759 pKa = 3.22TFTYY763 pKa = 9.12TVNGGSTGTVSVMVTCVDD781 pKa = 4.17DD782 pKa = 5.0LPVAVDD788 pKa = 4.07DD789 pKa = 4.85SEE791 pKa = 4.83QVTLNATATALMVLANDD808 pKa = 3.74TDD810 pKa = 3.73VDD812 pKa = 4.01GGALSIASVTQPTNGTVVITGGGTGLTYY840 pKa = 10.64EE841 pKa = 4.82PDD843 pKa = 3.11AGYY846 pKa = 10.96CDD848 pKa = 5.36DD849 pKa = 5.29PPGTTPDD856 pKa = 3.27TFTYY860 pKa = 9.17TLNGGSTAMVSVRR873 pKa = 11.84VVCDD877 pKa = 4.05DD878 pKa = 4.06PPVAVDD884 pKa = 3.52DD885 pKa = 4.57AKK887 pKa = 10.56TVVEE891 pKa = 4.71DD892 pKa = 3.93AAATAVDD899 pKa = 3.93VLANDD904 pKa = 3.87TDD906 pKa = 3.85VDD908 pKa = 3.88GGPISIASATQPTNGTVVITGGGTGLTYY936 pKa = 10.67QPDD939 pKa = 3.23ASYY942 pKa = 11.5CNNPPGTTLDD952 pKa = 3.47TFTYY956 pKa = 9.33TLTPGGSTATVSVTVTCVDD975 pKa = 3.83NLPVAVDD982 pKa = 3.58DD983 pKa = 4.63AKK985 pKa = 10.56TVVEE989 pKa = 4.71DD990 pKa = 3.93AAATAVDD997 pKa = 3.93VLANDD1002 pKa = 3.87TDD1004 pKa = 3.85VDD1006 pKa = 3.88GGPISIASVNQPTNGTVVISGGGTGLTYY1034 pKa = 10.68QPDD1037 pKa = 3.6ANYY1040 pKa = 10.66CNNPPGTTLDD1050 pKa = 3.47TFTYY1054 pKa = 9.33TLTPGGSTATVSLTVTCVNDD1074 pKa = 3.61PPNAVDD1080 pKa = 4.07DD1081 pKa = 4.09ATSVGEE1087 pKa = 4.36GAAGAAVPVLANDD1100 pKa = 3.94TDD1102 pKa = 4.45PDD1104 pKa = 3.95PTDD1107 pKa = 3.43VLSVTSVDD1115 pKa = 3.39TTGTIGGVSLVGGVVTYY1132 pKa = 10.87NPNGQFEE1139 pKa = 4.48YY1140 pKa = 10.65LAAGATATDD1149 pKa = 3.72TFSYY1153 pKa = 9.86TVSDD1157 pKa = 4.24GNGGTDD1163 pKa = 3.28TAVVTVTVTGANDD1176 pKa = 3.62PLVAVNDD1183 pKa = 3.94TGTTDD1188 pKa = 3.3EE1189 pKa = 5.06DD1190 pKa = 4.14TTLSQSAPGVLANDD1204 pKa = 4.02TDD1206 pKa = 4.75PDD1208 pKa = 3.85TGDD1211 pKa = 3.07TRR1213 pKa = 11.84TVTRR1217 pKa = 11.84LNGSATLTGTSTSGAAVSIAANGSFTYY1244 pKa = 10.7NPGGLFQGLSTGQSATDD1261 pKa = 3.41TFTYY1265 pKa = 9.08TAQDD1269 pKa = 3.29GSGAQSTATVTITINGVSDD1288 pKa = 3.76APTAVTDD1295 pKa = 3.81SFQTHH1300 pKa = 6.31GNTNLFVGTSRR1311 pKa = 11.84PVGEE1315 pKa = 4.4AGKK1318 pKa = 10.44VLTGSVLTNDD1328 pKa = 4.13TDD1330 pKa = 4.56PDD1332 pKa = 3.86TAQANLVAEE1341 pKa = 4.58PVTNAPTTLGGSITIEE1357 pKa = 3.61SDD1359 pKa = 3.62GNFTFYY1365 pKa = 10.47PDD1367 pKa = 4.81DD1368 pKa = 4.9GDD1370 pKa = 4.49SGVTDD1375 pKa = 3.41SFTYY1379 pKa = 9.9RR1380 pKa = 11.84VCDD1383 pKa = 4.02ASPCNASTVANATGTLNLPISSSQVWYY1410 pKa = 10.49VRR1412 pKa = 11.84NNAVAGGDD1420 pKa = 3.9GTSEE1424 pKa = 4.46APFDD1428 pKa = 3.93TLGEE1432 pKa = 4.27AEE1434 pKa = 4.25TAADD1438 pKa = 4.6AGDD1441 pKa = 3.52TTYY1444 pKa = 11.17VFDD1447 pKa = 5.75GDD1449 pKa = 3.88NTSLNLGSGFVMAAGEE1465 pKa = 4.19RR1466 pKa = 11.84LLGEE1470 pKa = 4.26AVTLQVGADD1479 pKa = 3.47VLYY1482 pKa = 10.47TGVPANRR1489 pKa = 11.84PTLTANDD1496 pKa = 3.84EE1497 pKa = 4.45DD1498 pKa = 4.9VVTLANATTVRR1509 pKa = 11.84GVQLDD1514 pKa = 3.73PQGAGGGIAGIGTASGPGTSTIADD1538 pKa = 3.29VRR1540 pKa = 11.84IIDD1543 pKa = 3.72TGTAGTQPGLEE1554 pKa = 4.05LDD1556 pKa = 4.17GVEE1559 pKa = 4.12GTYY1562 pKa = 10.34DD1563 pKa = 3.38ISDD1566 pKa = 3.73LTVDD1570 pKa = 3.73NSAATGQTAGSIGVRR1585 pKa = 11.84LNRR1588 pKa = 11.84NILGFTANFASAGTISLTTKK1608 pKa = 9.6GAKK1611 pKa = 9.82ALEE1614 pKa = 4.11VTGANMGLASVFDD1627 pKa = 5.71DD1628 pKa = 3.29ITVTGSPTGGVSMVNSLGSGTAFGDD1653 pKa = 4.11GVGVDD1658 pKa = 4.18LALTTTGGSAFNLQGTGTVSVPAAGTANVNATAGAGVFVNGSSGSSLAFDD1708 pKa = 4.26DD1709 pKa = 4.0VTVVSNPSGVGIAAVALGTGSFSAATGSISNTAGTAFTTGTGNGPISYY1757 pKa = 10.36GGTINNGSGRR1767 pKa = 11.84SVEE1770 pKa = 4.13VSARR1774 pKa = 11.84TGGTVTLTGNINDD1787 pKa = 4.15TNDD1790 pKa = 2.99VGGGIFVTGNSGGSTVLSGTTKK1812 pKa = 10.05TLNTGSEE1819 pKa = 4.2KK1820 pKa = 10.85AVDD1823 pKa = 4.58FSSSDD1828 pKa = 3.06GHH1830 pKa = 6.03TLTLSGGGLDD1840 pKa = 3.52IDD1842 pKa = 4.38TVSGTGVDD1850 pKa = 3.59AASSGTLNITGAGNTIDD1867 pKa = 3.64TSAGKK1872 pKa = 10.02ALSVSDD1878 pKa = 3.89TDD1880 pKa = 3.88VGPSPLSLQRR1890 pKa = 11.84ISSLGGAGITLSNTGSNNALVVASSGSGTCTAADD1924 pKa = 3.35QSGCTGGEE1932 pKa = 3.62IRR1934 pKa = 11.84SATGVDD1940 pKa = 3.81DD1941 pKa = 5.41SGALPTGTGIVLNNTRR1957 pKa = 11.84GVSLTRR1963 pKa = 11.84MHH1965 pKa = 6.8LHH1967 pKa = 6.38DD1968 pKa = 4.6HH1969 pKa = 6.11SNYY1972 pKa = 9.72GIRR1975 pKa = 11.84GTSVVGFSMVDD1986 pKa = 3.26SVVNGVNGTNGTTTFKK2002 pKa = 10.73DD2003 pKa = 3.62GSVRR2007 pKa = 11.84FVEE2010 pKa = 4.34LTGTVAMTNVALSGGYY2026 pKa = 7.55FTNLMVDD2033 pKa = 3.55NTAGILNGTFDD2044 pKa = 4.19NVDD2047 pKa = 3.37SGTINATGGDD2057 pKa = 3.66DD2058 pKa = 3.5AVQFEE2063 pKa = 5.29GIGTSTMNVDD2073 pKa = 4.1YY2074 pKa = 10.45KK2075 pKa = 11.32NSAIGTATGDD2085 pKa = 3.54LFQYY2089 pKa = 10.42IGDD2092 pKa = 3.94GTGGGNLDD2100 pKa = 3.54LTSNAFTNNEE2110 pKa = 3.75PSIATGGGGVAIVAGAKK2127 pKa = 9.64GAATLDD2133 pKa = 3.29IVSNTFRR2140 pKa = 11.84DD2141 pKa = 3.76SLTNALTVIKK2151 pKa = 10.57SADD2154 pKa = 3.3ATAGTNPLVSNITGNTIGVAATANSGSFEE2183 pKa = 4.23GDD2185 pKa = 2.72GMEE2188 pKa = 3.91ITTFGDD2194 pKa = 3.54GNATFNVLNNNISQYY2209 pKa = 10.39NSSGIQFVAGSGVTATGQMNLNVSGNVVGNPGTNPLITLLQGIRR2253 pKa = 11.84VDD2255 pKa = 4.04SGVDD2259 pKa = 3.15TADD2262 pKa = 3.41TFSTCVKK2269 pKa = 10.2FGANSISGAGGSAGRR2284 pKa = 11.84DD2285 pKa = 3.59FRR2287 pKa = 11.84LVASQQTTLRR2297 pKa = 11.84QPGYY2301 pKa = 10.82VGGSTDD2307 pKa = 2.99GTAFANYY2314 pKa = 8.74AASLIGSGAAGTAVANAPGTFSGAGSTCPP2343 pKa = 3.86
Molecular weight: 230.53 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A3THR1|A3THR1_9MICO Putative transcriptional regulator OS=Janibacter sp. HTCC2649 OX=313589 GN=JNB_09369 PE=4 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
Molecular weight: 4.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1293981 |
18 |
3687 |
317.1 |
33.84 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.954 ± 0.052 | 0.658 ± 0.008 |
6.333 ± 0.035 | 5.438 ± 0.04 |
2.834 ± 0.026 | 9.291 ± 0.041 |
2.221 ± 0.022 | 3.995 ± 0.028 |
2.312 ± 0.029 | 9.919 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.892 ± 0.016 | 1.979 ± 0.025 |
5.459 ± 0.029 | 2.8 ± 0.018 |
7.035 ± 0.045 | 5.782 ± 0.026 |
6.556 ± 0.04 | 9.156 ± 0.037 |
1.573 ± 0.017 | 1.814 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
