
Serpula lacrymans var. lacrymans (strain S7.3) (Dry rot fungus)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota; Agaricomycotina; Agaricomycetes; Agaricomycetidae; Boletales; Coniophorineae; Serpulaceae; Serpula; Serpula lacrymans; Serpula lacrymans var. lacrymans
Average proteome isoelectric point is 6.77
Get precalculated fractions of proteins
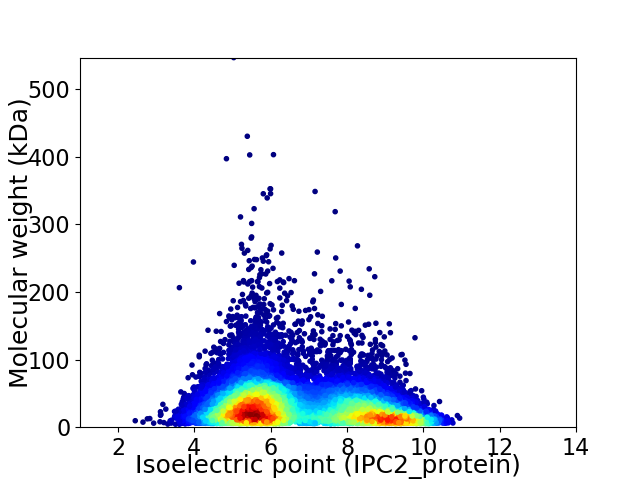
Virtual 2D-PAGE plot for 14339 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F8Q7Z7|F8Q7Z7_SERL3 Zn(2)-C6 fungal-type domain-containing protein OS=Serpula lacrymans var. lacrymans (strain S7.3) OX=936435 GN=SERLA73DRAFT_59903 PE=4 SV=1
MM1 pKa = 6.86STEE4 pKa = 4.22SFDD7 pKa = 4.1FLHH10 pKa = 7.08LARR13 pKa = 11.84SKK15 pKa = 10.88LHH17 pKa = 5.41TAIGGKK23 pKa = 9.17DD24 pKa = 3.12NCSLHH29 pKa = 6.6RR30 pKa = 11.84WVLLKK35 pKa = 10.93NSIVRR40 pKa = 11.84TPPKK44 pKa = 10.39DD45 pKa = 3.77GEE47 pKa = 4.67VQNPMPDD54 pKa = 3.39VNSVYY59 pKa = 11.02SFDD62 pKa = 5.53DD63 pKa = 3.75EE64 pKa = 6.21GDD66 pKa = 3.75GDD68 pKa = 4.76DD69 pKa = 4.52EE70 pKa = 6.6DD71 pKa = 6.48DD72 pKa = 4.24GVEE75 pKa = 4.0EE76 pKa = 5.04EE77 pKa = 4.72IDD79 pKa = 3.52SFMFPDD85 pKa = 4.02ASNLVDD91 pKa = 4.98CGGADD96 pKa = 3.6VNASEE101 pKa = 5.64AEE103 pKa = 4.08WLDD106 pKa = 3.61SLLEE110 pKa = 4.03TLGEE114 pKa = 4.43DD115 pKa = 5.08GEE117 pKa = 4.67DD118 pKa = 3.34LEE120 pKa = 4.5QIRR123 pKa = 11.84VSVLPVEE130 pKa = 5.08DD131 pKa = 5.91DD132 pKa = 3.96EE133 pKa = 5.98DD134 pKa = 4.37LQTSPLVSPMSSSDD148 pKa = 3.74DD149 pKa = 3.86LLNQLAYY156 pKa = 9.73YY157 pKa = 9.22SPPIAVPYY165 pKa = 8.01PVPYY169 pKa = 10.28PPFHH173 pKa = 7.16PPLIRR178 pKa = 11.84PCEE181 pKa = 4.17LGPIIDD187 pKa = 4.37SPLLSSLPAYY197 pKa = 9.88DD198 pKa = 5.69DD199 pKa = 4.04PLPYY203 pKa = 10.4YY204 pKa = 9.98DD205 pKa = 4.26TDD207 pKa = 4.23EE208 pKa = 5.73IDD210 pKa = 4.47DD211 pKa = 4.66LSVPDD216 pKa = 5.32AIEE219 pKa = 4.35DD220 pKa = 3.92TSDD223 pKa = 3.9DD224 pKa = 3.83EE225 pKa = 5.31SDD227 pKa = 3.6IPSTPSLVRR236 pKa = 11.84SSTQSFVDD244 pKa = 3.92PASVPLPAEE253 pKa = 3.93RR254 pKa = 11.84RR255 pKa = 11.84RR256 pKa = 11.84QRR258 pKa = 11.84MQPHH262 pKa = 6.15VYY264 pKa = 9.57IGTMDD269 pKa = 3.98PYY271 pKa = 10.73FYY273 pKa = 10.4PFEE276 pKa = 5.39LDD278 pKa = 3.61PLPFPDD284 pKa = 3.52EE285 pKa = 4.62DD286 pKa = 4.16RR287 pKa = 11.84FASYY291 pKa = 11.23NPIYY295 pKa = 10.42SEE297 pKa = 4.38CC298 pKa = 3.85
MM1 pKa = 6.86STEE4 pKa = 4.22SFDD7 pKa = 4.1FLHH10 pKa = 7.08LARR13 pKa = 11.84SKK15 pKa = 10.88LHH17 pKa = 5.41TAIGGKK23 pKa = 9.17DD24 pKa = 3.12NCSLHH29 pKa = 6.6RR30 pKa = 11.84WVLLKK35 pKa = 10.93NSIVRR40 pKa = 11.84TPPKK44 pKa = 10.39DD45 pKa = 3.77GEE47 pKa = 4.67VQNPMPDD54 pKa = 3.39VNSVYY59 pKa = 11.02SFDD62 pKa = 5.53DD63 pKa = 3.75EE64 pKa = 6.21GDD66 pKa = 3.75GDD68 pKa = 4.76DD69 pKa = 4.52EE70 pKa = 6.6DD71 pKa = 6.48DD72 pKa = 4.24GVEE75 pKa = 4.0EE76 pKa = 5.04EE77 pKa = 4.72IDD79 pKa = 3.52SFMFPDD85 pKa = 4.02ASNLVDD91 pKa = 4.98CGGADD96 pKa = 3.6VNASEE101 pKa = 5.64AEE103 pKa = 4.08WLDD106 pKa = 3.61SLLEE110 pKa = 4.03TLGEE114 pKa = 4.43DD115 pKa = 5.08GEE117 pKa = 4.67DD118 pKa = 3.34LEE120 pKa = 4.5QIRR123 pKa = 11.84VSVLPVEE130 pKa = 5.08DD131 pKa = 5.91DD132 pKa = 3.96EE133 pKa = 5.98DD134 pKa = 4.37LQTSPLVSPMSSSDD148 pKa = 3.74DD149 pKa = 3.86LLNQLAYY156 pKa = 9.73YY157 pKa = 9.22SPPIAVPYY165 pKa = 8.01PVPYY169 pKa = 10.28PPFHH173 pKa = 7.16PPLIRR178 pKa = 11.84PCEE181 pKa = 4.17LGPIIDD187 pKa = 4.37SPLLSSLPAYY197 pKa = 9.88DD198 pKa = 5.69DD199 pKa = 4.04PLPYY203 pKa = 10.4YY204 pKa = 9.98DD205 pKa = 4.26TDD207 pKa = 4.23EE208 pKa = 5.73IDD210 pKa = 4.47DD211 pKa = 4.66LSVPDD216 pKa = 5.32AIEE219 pKa = 4.35DD220 pKa = 3.92TSDD223 pKa = 3.9DD224 pKa = 3.83EE225 pKa = 5.31SDD227 pKa = 3.6IPSTPSLVRR236 pKa = 11.84SSTQSFVDD244 pKa = 3.92PASVPLPAEE253 pKa = 3.93RR254 pKa = 11.84RR255 pKa = 11.84RR256 pKa = 11.84QRR258 pKa = 11.84MQPHH262 pKa = 6.15VYY264 pKa = 9.57IGTMDD269 pKa = 3.98PYY271 pKa = 10.73FYY273 pKa = 10.4PFEE276 pKa = 5.39LDD278 pKa = 3.61PLPFPDD284 pKa = 3.52EE285 pKa = 4.62DD286 pKa = 4.16RR287 pKa = 11.84FASYY291 pKa = 11.23NPIYY295 pKa = 10.42SEE297 pKa = 4.38CC298 pKa = 3.85
Molecular weight: 33.17 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F8PVY8|F8PVY8_SERL3 Uncharacterized protein OS=Serpula lacrymans var. lacrymans (strain S7.3) OX=936435 GN=SERLA73DRAFT_106193 PE=4 SV=1
MM1 pKa = 7.52PRR3 pKa = 11.84ILRR6 pKa = 11.84QLVRR10 pKa = 11.84LLSRR14 pKa = 11.84PPTLQALPSRR24 pKa = 11.84AASHH28 pKa = 6.46LRR30 pKa = 11.84AFQATLPFCSQILNNHH46 pKa = 5.78NPLLFGPAQHH56 pKa = 6.97RR57 pKa = 11.84ISSPILAAVHH67 pKa = 4.82QVRR70 pKa = 11.84FAARR74 pKa = 11.84GTEE77 pKa = 4.06YY78 pKa = 10.7QPSQRR83 pKa = 11.84KK84 pKa = 8.85RR85 pKa = 11.84KK86 pKa = 9.17RR87 pKa = 11.84KK88 pKa = 9.56HH89 pKa = 5.55GFLARR94 pKa = 11.84KK95 pKa = 9.69RR96 pKa = 11.84SITGRR101 pKa = 11.84RR102 pKa = 11.84ILARR106 pKa = 11.84RR107 pKa = 11.84KK108 pKa = 9.31ARR110 pKa = 11.84GRR112 pKa = 11.84TFLSHH117 pKa = 6.99
MM1 pKa = 7.52PRR3 pKa = 11.84ILRR6 pKa = 11.84QLVRR10 pKa = 11.84LLSRR14 pKa = 11.84PPTLQALPSRR24 pKa = 11.84AASHH28 pKa = 6.46LRR30 pKa = 11.84AFQATLPFCSQILNNHH46 pKa = 5.78NPLLFGPAQHH56 pKa = 6.97RR57 pKa = 11.84ISSPILAAVHH67 pKa = 4.82QVRR70 pKa = 11.84FAARR74 pKa = 11.84GTEE77 pKa = 4.06YY78 pKa = 10.7QPSQRR83 pKa = 11.84KK84 pKa = 8.85RR85 pKa = 11.84KK86 pKa = 9.17RR87 pKa = 11.84KK88 pKa = 9.56HH89 pKa = 5.55GFLARR94 pKa = 11.84KK95 pKa = 9.69RR96 pKa = 11.84SITGRR101 pKa = 11.84RR102 pKa = 11.84ILARR106 pKa = 11.84RR107 pKa = 11.84KK108 pKa = 9.31ARR110 pKa = 11.84GRR112 pKa = 11.84TFLSHH117 pKa = 6.99
Molecular weight: 13.43 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4645570 |
42 |
4889 |
324.0 |
36.0 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.881 ± 0.019 | 1.372 ± 0.008 |
5.523 ± 0.016 | 5.816 ± 0.021 |
3.836 ± 0.013 | 6.352 ± 0.021 |
2.674 ± 0.012 | 5.269 ± 0.015 |
4.913 ± 0.024 | 9.298 ± 0.024 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.23 ± 0.008 | 3.766 ± 0.012 |
5.952 ± 0.024 | 3.928 ± 0.014 |
5.825 ± 0.021 | 8.72 ± 0.026 |
5.947 ± 0.015 | 6.367 ± 0.016 |
1.491 ± 0.008 | 2.841 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
