
Halanaerobium saccharolyticum subsp. saccharolyticum DSM 6643
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Halanaerobiales; Halanaerobiaceae; Halanaerobium; Halanaerobium saccharolyticum; Halanaerobium saccharolyticum subsp. saccharolyticum
Average proteome isoelectric point is 6.13
Get precalculated fractions of proteins
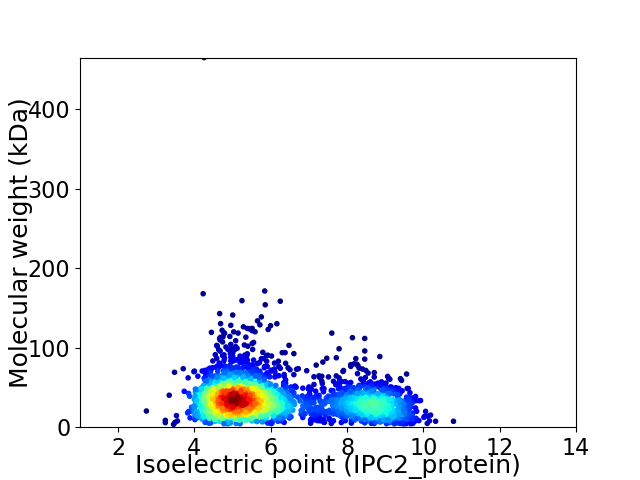
Virtual 2D-PAGE plot for 2662 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
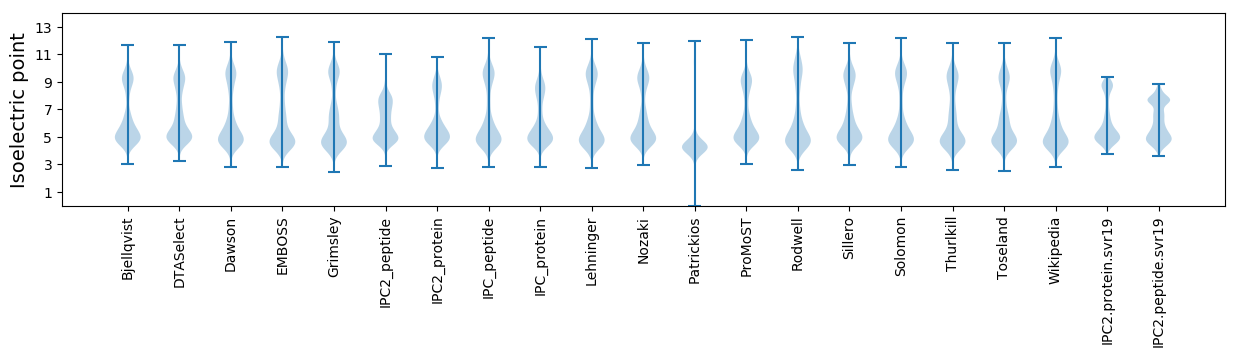
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|M5EGC9|M5EGC9_9FIRM Late competence protein ComEA DNA receptor OS=Halanaerobium saccharolyticum subsp. saccharolyticum DSM 6643 OX=1293054 GN=HSACCH_02065 PE=4 SV=1
MM1 pKa = 7.59KK2 pKa = 10.49KK3 pKa = 9.03LTITIALMLVVALAMPAFGASFSDD27 pKa = 3.87VPSDD31 pKa = 2.75HH32 pKa = 6.79WAYY35 pKa = 11.04NAINKK40 pKa = 8.94LVAAGIVEE48 pKa = 4.82GYY50 pKa = 9.83PDD52 pKa = 4.23GEE54 pKa = 4.41YY55 pKa = 10.38KK56 pKa = 10.64GQQSMTRR63 pKa = 11.84YY64 pKa = 8.92EE65 pKa = 4.09MAVMVSRR72 pKa = 11.84ALDD75 pKa = 3.94NIVDD79 pKa = 3.7EE80 pKa = 4.47MEE82 pKa = 4.32AMGEE86 pKa = 4.14GLTTGQAEE94 pKa = 4.49DD95 pKa = 3.17VTAIVKK101 pKa = 10.45SLMEE105 pKa = 4.62KK106 pKa = 8.88NTNDD110 pKa = 3.79EE111 pKa = 4.3LSDD114 pKa = 3.63AQAEE118 pKa = 4.25EE119 pKa = 4.16VADD122 pKa = 3.71IVDD125 pKa = 3.58ALTFEE130 pKa = 4.75LRR132 pKa = 11.84SEE134 pKa = 4.25LKK136 pKa = 10.78VLGAEE141 pKa = 3.96VDD143 pKa = 3.87TLGKK147 pKa = 10.44DD148 pKa = 3.53VDD150 pKa = 3.77EE151 pKa = 4.87LAAKK155 pKa = 9.63VEE157 pKa = 3.99AMEE160 pKa = 4.24VPEE163 pKa = 4.67DD164 pKa = 3.97NIEE167 pKa = 4.06FGMDD171 pKa = 2.95VTTTFEE177 pKa = 3.86AASYY181 pKa = 11.17GEE183 pKa = 4.31DD184 pKa = 3.45AAEE187 pKa = 3.93EE188 pKa = 5.43GYY190 pKa = 9.23TINLLEE196 pKa = 5.59DD197 pKa = 3.85GDD199 pKa = 5.37AIDD202 pKa = 5.26EE203 pKa = 4.77SFTSDD208 pKa = 3.41PDD210 pKa = 3.3EE211 pKa = 4.56FTSKK215 pKa = 10.64KK216 pKa = 10.78AFFQEE221 pKa = 3.54YY222 pKa = 10.16DD223 pKa = 3.54FNINGMLGDD232 pKa = 3.72ATFNLDD238 pKa = 2.52VDD240 pKa = 4.61TIANVFTEE248 pKa = 4.25EE249 pKa = 3.97DD250 pKa = 3.43HH251 pKa = 6.27VANAVSEE258 pKa = 4.4TNQNDD263 pKa = 3.17FMMDD267 pKa = 3.25SALLEE272 pKa = 4.04VSYY275 pKa = 11.43NNYY278 pKa = 9.94SFQIGDD284 pKa = 3.77LNDD287 pKa = 3.54YY288 pKa = 10.91SVAPYY293 pKa = 10.57FNDD296 pKa = 3.24EE297 pKa = 4.06EE298 pKa = 4.19DD299 pKa = 3.88RR300 pKa = 11.84EE301 pKa = 4.62GIEE304 pKa = 3.96MTTSYY309 pKa = 10.19MDD311 pKa = 3.66NDD313 pKa = 2.93IKK315 pKa = 10.48TFVLGADD322 pKa = 3.43VSEE325 pKa = 4.52DD326 pKa = 3.5TFGTTTVDD334 pKa = 2.69SDD336 pKa = 4.34ANDD339 pKa = 3.27EE340 pKa = 4.17EE341 pKa = 5.58YY342 pKa = 11.3YY343 pKa = 10.62GVEE346 pKa = 4.11VARR349 pKa = 11.84DD350 pKa = 3.58MEE352 pKa = 4.69FGRR355 pKa = 11.84VTGKK359 pKa = 10.49LYY361 pKa = 10.35HH362 pKa = 6.8ARR364 pKa = 11.84DD365 pKa = 3.21IDD367 pKa = 3.81FDD369 pKa = 4.07GFNMAEE375 pKa = 4.14YY376 pKa = 10.68AVTAEE381 pKa = 4.27KK382 pKa = 11.24NEE384 pKa = 4.16TTILAAQIEE393 pKa = 4.76DD394 pKa = 3.83VVITDD399 pKa = 5.36AISMGAEE406 pKa = 3.95VAFSDD411 pKa = 4.11WEE413 pKa = 3.9RR414 pKa = 11.84DD415 pKa = 3.07AYY417 pKa = 7.82TTYY420 pKa = 11.33DD421 pKa = 3.29SDD423 pKa = 3.88ASATEE428 pKa = 4.17AVAADD433 pKa = 4.78DD434 pKa = 5.77DD435 pKa = 4.69SDD437 pKa = 4.19IYY439 pKa = 11.47FNVNGEE445 pKa = 4.22FVATEE450 pKa = 4.32EE451 pKa = 4.33LTLDD455 pKa = 3.45ATIEE459 pKa = 4.33TVGEE463 pKa = 4.32DD464 pKa = 3.97FVGPYY469 pKa = 10.29NDD471 pKa = 4.67LEE473 pKa = 4.24EE474 pKa = 5.74ASDD477 pKa = 3.84YY478 pKa = 12.04DD479 pKa = 3.86MFNVGAEE486 pKa = 4.1YY487 pKa = 11.01VLNEE491 pKa = 3.99NNTVTGAYY499 pKa = 9.14TLVDD503 pKa = 3.63HH504 pKa = 7.02NEE506 pKa = 4.27PEE508 pKa = 4.42DD509 pKa = 3.86KK510 pKa = 10.53STVEE514 pKa = 4.18LGLEE518 pKa = 3.89NVYY521 pKa = 11.43GDD523 pKa = 3.8FTNNASVEE531 pKa = 4.19FVMNDD536 pKa = 3.74DD537 pKa = 3.71YY538 pKa = 11.87TDD540 pKa = 4.24DD541 pKa = 4.48YY542 pKa = 10.47EE543 pKa = 4.49TMIIVLGTEE552 pKa = 4.32YY553 pKa = 10.95AWDD556 pKa = 3.55EE557 pKa = 4.25TLTLGAEE564 pKa = 4.41LTNKK568 pKa = 9.28TKK570 pKa = 11.08DD571 pKa = 3.41EE572 pKa = 4.84AGTDD576 pKa = 3.79AITYY580 pKa = 10.08NYY582 pKa = 8.15LTAFADD588 pKa = 4.82KK589 pKa = 10.42EE590 pKa = 4.07LADD593 pKa = 4.27NITWNTEE600 pKa = 3.24AFFIDD605 pKa = 4.56GEE607 pKa = 4.15LDD609 pKa = 3.87EE610 pKa = 5.65INEE613 pKa = 4.23GQGNGMTTSLSVSFF627 pKa = 4.81
MM1 pKa = 7.59KK2 pKa = 10.49KK3 pKa = 9.03LTITIALMLVVALAMPAFGASFSDD27 pKa = 3.87VPSDD31 pKa = 2.75HH32 pKa = 6.79WAYY35 pKa = 11.04NAINKK40 pKa = 8.94LVAAGIVEE48 pKa = 4.82GYY50 pKa = 9.83PDD52 pKa = 4.23GEE54 pKa = 4.41YY55 pKa = 10.38KK56 pKa = 10.64GQQSMTRR63 pKa = 11.84YY64 pKa = 8.92EE65 pKa = 4.09MAVMVSRR72 pKa = 11.84ALDD75 pKa = 3.94NIVDD79 pKa = 3.7EE80 pKa = 4.47MEE82 pKa = 4.32AMGEE86 pKa = 4.14GLTTGQAEE94 pKa = 4.49DD95 pKa = 3.17VTAIVKK101 pKa = 10.45SLMEE105 pKa = 4.62KK106 pKa = 8.88NTNDD110 pKa = 3.79EE111 pKa = 4.3LSDD114 pKa = 3.63AQAEE118 pKa = 4.25EE119 pKa = 4.16VADD122 pKa = 3.71IVDD125 pKa = 3.58ALTFEE130 pKa = 4.75LRR132 pKa = 11.84SEE134 pKa = 4.25LKK136 pKa = 10.78VLGAEE141 pKa = 3.96VDD143 pKa = 3.87TLGKK147 pKa = 10.44DD148 pKa = 3.53VDD150 pKa = 3.77EE151 pKa = 4.87LAAKK155 pKa = 9.63VEE157 pKa = 3.99AMEE160 pKa = 4.24VPEE163 pKa = 4.67DD164 pKa = 3.97NIEE167 pKa = 4.06FGMDD171 pKa = 2.95VTTTFEE177 pKa = 3.86AASYY181 pKa = 11.17GEE183 pKa = 4.31DD184 pKa = 3.45AAEE187 pKa = 3.93EE188 pKa = 5.43GYY190 pKa = 9.23TINLLEE196 pKa = 5.59DD197 pKa = 3.85GDD199 pKa = 5.37AIDD202 pKa = 5.26EE203 pKa = 4.77SFTSDD208 pKa = 3.41PDD210 pKa = 3.3EE211 pKa = 4.56FTSKK215 pKa = 10.64KK216 pKa = 10.78AFFQEE221 pKa = 3.54YY222 pKa = 10.16DD223 pKa = 3.54FNINGMLGDD232 pKa = 3.72ATFNLDD238 pKa = 2.52VDD240 pKa = 4.61TIANVFTEE248 pKa = 4.25EE249 pKa = 3.97DD250 pKa = 3.43HH251 pKa = 6.27VANAVSEE258 pKa = 4.4TNQNDD263 pKa = 3.17FMMDD267 pKa = 3.25SALLEE272 pKa = 4.04VSYY275 pKa = 11.43NNYY278 pKa = 9.94SFQIGDD284 pKa = 3.77LNDD287 pKa = 3.54YY288 pKa = 10.91SVAPYY293 pKa = 10.57FNDD296 pKa = 3.24EE297 pKa = 4.06EE298 pKa = 4.19DD299 pKa = 3.88RR300 pKa = 11.84EE301 pKa = 4.62GIEE304 pKa = 3.96MTTSYY309 pKa = 10.19MDD311 pKa = 3.66NDD313 pKa = 2.93IKK315 pKa = 10.48TFVLGADD322 pKa = 3.43VSEE325 pKa = 4.52DD326 pKa = 3.5TFGTTTVDD334 pKa = 2.69SDD336 pKa = 4.34ANDD339 pKa = 3.27EE340 pKa = 4.17EE341 pKa = 5.58YY342 pKa = 11.3YY343 pKa = 10.62GVEE346 pKa = 4.11VARR349 pKa = 11.84DD350 pKa = 3.58MEE352 pKa = 4.69FGRR355 pKa = 11.84VTGKK359 pKa = 10.49LYY361 pKa = 10.35HH362 pKa = 6.8ARR364 pKa = 11.84DD365 pKa = 3.21IDD367 pKa = 3.81FDD369 pKa = 4.07GFNMAEE375 pKa = 4.14YY376 pKa = 10.68AVTAEE381 pKa = 4.27KK382 pKa = 11.24NEE384 pKa = 4.16TTILAAQIEE393 pKa = 4.76DD394 pKa = 3.83VVITDD399 pKa = 5.36AISMGAEE406 pKa = 3.95VAFSDD411 pKa = 4.11WEE413 pKa = 3.9RR414 pKa = 11.84DD415 pKa = 3.07AYY417 pKa = 7.82TTYY420 pKa = 11.33DD421 pKa = 3.29SDD423 pKa = 3.88ASATEE428 pKa = 4.17AVAADD433 pKa = 4.78DD434 pKa = 5.77DD435 pKa = 4.69SDD437 pKa = 4.19IYY439 pKa = 11.47FNVNGEE445 pKa = 4.22FVATEE450 pKa = 4.32EE451 pKa = 4.33LTLDD455 pKa = 3.45ATIEE459 pKa = 4.33TVGEE463 pKa = 4.32DD464 pKa = 3.97FVGPYY469 pKa = 10.29NDD471 pKa = 4.67LEE473 pKa = 4.24EE474 pKa = 5.74ASDD477 pKa = 3.84YY478 pKa = 12.04DD479 pKa = 3.86MFNVGAEE486 pKa = 4.1YY487 pKa = 11.01VLNEE491 pKa = 3.99NNTVTGAYY499 pKa = 9.14TLVDD503 pKa = 3.63HH504 pKa = 7.02NEE506 pKa = 4.27PEE508 pKa = 4.42DD509 pKa = 3.86KK510 pKa = 10.53STVEE514 pKa = 4.18LGLEE518 pKa = 3.89NVYY521 pKa = 11.43GDD523 pKa = 3.8FTNNASVEE531 pKa = 4.19FVMNDD536 pKa = 3.74DD537 pKa = 3.71YY538 pKa = 11.87TDD540 pKa = 4.24DD541 pKa = 4.48YY542 pKa = 10.47EE543 pKa = 4.49TMIIVLGTEE552 pKa = 4.32YY553 pKa = 10.95AWDD556 pKa = 3.55EE557 pKa = 4.25TLTLGAEE564 pKa = 4.41LTNKK568 pKa = 9.28TKK570 pKa = 11.08DD571 pKa = 3.41EE572 pKa = 4.84AGTDD576 pKa = 3.79AITYY580 pKa = 10.08NYY582 pKa = 8.15LTAFADD588 pKa = 4.82KK589 pKa = 10.42EE590 pKa = 4.07LADD593 pKa = 4.27NITWNTEE600 pKa = 3.24AFFIDD605 pKa = 4.56GEE607 pKa = 4.15LDD609 pKa = 3.87EE610 pKa = 5.65INEE613 pKa = 4.23GQGNGMTTSLSVSFF627 pKa = 4.81
Molecular weight: 69.06 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|M5EF68|M5EF68_9FIRM Uncharacterized protein OS=Halanaerobium saccharolyticum subsp. saccharolyticum DSM 6643 OX=1293054 GN=HSACCH_01585 PE=4 SV=1
MM1 pKa = 7.4NSKK4 pKa = 10.32KK5 pKa = 10.81GIIFTGGIIGILGGLLVVFGNPANMGICVACFIRR39 pKa = 11.84DD40 pKa = 3.12IAGAVGLHH48 pKa = 6.23RR49 pKa = 11.84AAVVQNIRR57 pKa = 11.84PEE59 pKa = 4.23IIGFILGAFLIARR72 pKa = 11.84STGEE76 pKa = 3.96FKK78 pKa = 11.21VKK80 pKa = 10.4GGSSPITRR88 pKa = 11.84FVIAMFVMIGALVFLGCPLRR108 pKa = 11.84MVLRR112 pKa = 11.84LAGGDD117 pKa = 3.23WNAIAGIIGFVLGVLAGSQLLKK139 pKa = 10.82KK140 pKa = 10.46GFTLGRR146 pKa = 11.84SYY148 pKa = 10.99KK149 pKa = 9.67INQINGYY156 pKa = 8.6ILPIIAVVLLIFRR169 pKa = 11.84IVRR172 pKa = 11.84PSFIIFSEE180 pKa = 4.1KK181 pKa = 10.78GPGASYY187 pKa = 11.02AFWAIALAAGIIIGILAQRR206 pKa = 11.84SRR208 pKa = 11.84ICMAGGIRR216 pKa = 11.84DD217 pKa = 4.16LYY219 pKa = 10.51LIKK222 pKa = 10.36DD223 pKa = 3.44SHH225 pKa = 7.26LITGFISIFIFAFIVNLIFGNFNPGFAGQPAAHH258 pKa = 6.33SQWLWNLLGMFLTGLGSVMLGGCPLRR284 pKa = 11.84QTILAGEE291 pKa = 4.69GNTDD295 pKa = 3.22SAVAFVGYY303 pKa = 10.07LVGAAISHH311 pKa = 5.82NFGLAGSGSGVSTNGRR327 pKa = 11.84IAVISGIVILFIIAYY342 pKa = 10.21SSIQFNKK349 pKa = 10.01KK350 pKa = 9.9AVV352 pKa = 3.22
MM1 pKa = 7.4NSKK4 pKa = 10.32KK5 pKa = 10.81GIIFTGGIIGILGGLLVVFGNPANMGICVACFIRR39 pKa = 11.84DD40 pKa = 3.12IAGAVGLHH48 pKa = 6.23RR49 pKa = 11.84AAVVQNIRR57 pKa = 11.84PEE59 pKa = 4.23IIGFILGAFLIARR72 pKa = 11.84STGEE76 pKa = 3.96FKK78 pKa = 11.21VKK80 pKa = 10.4GGSSPITRR88 pKa = 11.84FVIAMFVMIGALVFLGCPLRR108 pKa = 11.84MVLRR112 pKa = 11.84LAGGDD117 pKa = 3.23WNAIAGIIGFVLGVLAGSQLLKK139 pKa = 10.82KK140 pKa = 10.46GFTLGRR146 pKa = 11.84SYY148 pKa = 10.99KK149 pKa = 9.67INQINGYY156 pKa = 8.6ILPIIAVVLLIFRR169 pKa = 11.84IVRR172 pKa = 11.84PSFIIFSEE180 pKa = 4.1KK181 pKa = 10.78GPGASYY187 pKa = 11.02AFWAIALAAGIIIGILAQRR206 pKa = 11.84SRR208 pKa = 11.84ICMAGGIRR216 pKa = 11.84DD217 pKa = 4.16LYY219 pKa = 10.51LIKK222 pKa = 10.36DD223 pKa = 3.44SHH225 pKa = 7.26LITGFISIFIFAFIVNLIFGNFNPGFAGQPAAHH258 pKa = 6.33SQWLWNLLGMFLTGLGSVMLGGCPLRR284 pKa = 11.84QTILAGEE291 pKa = 4.69GNTDD295 pKa = 3.22SAVAFVGYY303 pKa = 10.07LVGAAISHH311 pKa = 5.82NFGLAGSGSGVSTNGRR327 pKa = 11.84IAVISGIVILFIIAYY342 pKa = 10.21SSIQFNKK349 pKa = 10.01KK350 pKa = 9.9AVV352 pKa = 3.22
Molecular weight: 37.0 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
861534 |
37 |
4311 |
323.6 |
36.43 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.134 ± 0.052 | 0.616 ± 0.014 |
5.776 ± 0.04 | 8.326 ± 0.06 |
4.494 ± 0.036 | 6.405 ± 0.049 |
1.459 ± 0.017 | 8.987 ± 0.049 |
8.286 ± 0.057 | 9.925 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.416 ± 0.024 | 5.722 ± 0.058 |
3.036 ± 0.025 | 3.038 ± 0.029 |
3.549 ± 0.033 | 6.02 ± 0.048 |
4.333 ± 0.028 | 6.034 ± 0.042 |
0.729 ± 0.015 | 3.715 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
