
Roseivivax marinus
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Roseobacteraceae; Roseivivax
Average proteome isoelectric point is 6.0
Get precalculated fractions of proteins
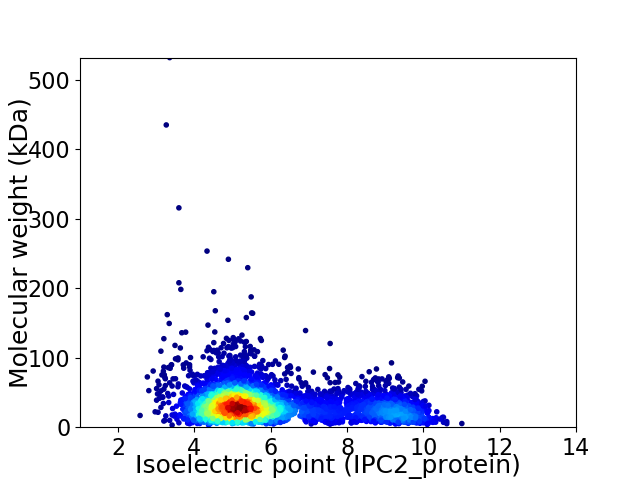
Virtual 2D-PAGE plot for 4264 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
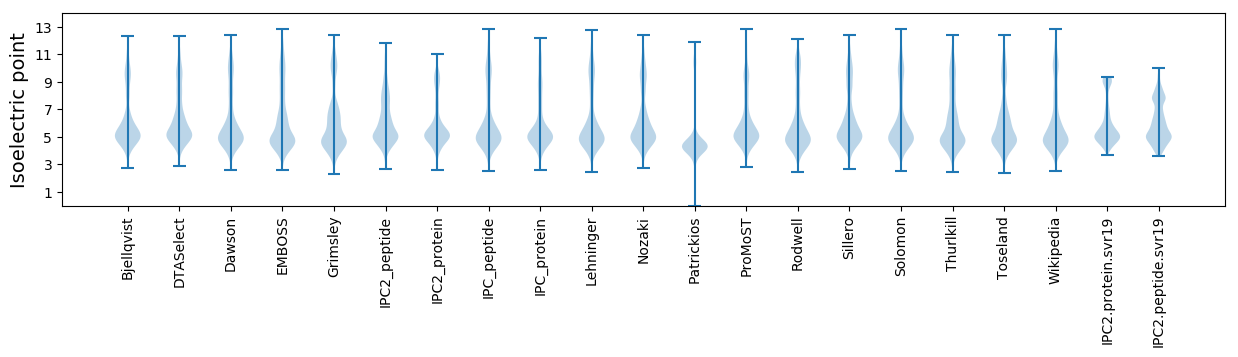
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|W4HNM1|W4HNM1_9RHOB Alpha-E domain-containing protein OS=Roseivivax marinus OX=1379903 GN=ATO8_00520 PE=4 SV=1
MM1 pKa = 7.22TVDD4 pKa = 3.67IYY6 pKa = 11.58APTSSDD12 pKa = 3.49SLSLTEE18 pKa = 4.29LDD20 pKa = 4.34LYY22 pKa = 10.76HH23 pKa = 7.01MIMDD27 pKa = 3.7YY28 pKa = 10.64RR29 pKa = 11.84AASGLPAIPLSQSLTTVAGRR49 pKa = 11.84HH50 pKa = 5.04TLDD53 pKa = 3.32SYY55 pKa = 12.0HH56 pKa = 6.93NIEE59 pKa = 4.19AAGVSLPSGYY69 pKa = 10.5SSPSWIHH76 pKa = 5.42SWSDD80 pKa = 2.94AYY82 pKa = 11.61YY83 pKa = 10.37DD84 pKa = 4.24GSDD87 pKa = 3.41SSVMRR92 pKa = 11.84DD93 pKa = 3.12APSRR97 pKa = 11.84LGTGYY102 pKa = 9.85PGGGYY107 pKa = 9.74EE108 pKa = 4.17ISVAGAQSIRR118 pKa = 11.84QTLDD122 pKa = 2.6MWIDD126 pKa = 3.88SPGHH130 pKa = 5.29NQLLVNLNSYY140 pKa = 11.34SDD142 pKa = 3.94DD143 pKa = 2.94TWNAIGVGVGFNAPVSGGSFTYY165 pKa = 10.83GNYY168 pKa = 8.37FHH170 pKa = 6.87VWFGKK175 pKa = 10.07VADD178 pKa = 4.05PAGPPVIDD186 pKa = 3.55GTNAGEE192 pKa = 4.47LIEE195 pKa = 4.26MTNFADD201 pKa = 3.99EE202 pKa = 4.14VRR204 pKa = 11.84DD205 pKa = 3.55RR206 pKa = 11.84GGNDD210 pKa = 3.04TVIGYY215 pKa = 9.49SGNDD219 pKa = 3.07TFYY222 pKa = 11.37VGGGANDD229 pKa = 3.28FRR231 pKa = 11.84GGDD234 pKa = 3.49GTDD237 pKa = 2.87RR238 pKa = 11.84AVIPATFAQVTATVDD253 pKa = 3.29GSVAQITLNGTTSTYY268 pKa = 10.72RR269 pKa = 11.84GIEE272 pKa = 3.77VFAFSDD278 pKa = 3.54GTTRR282 pKa = 11.84ALGQLNGGTSGGDD295 pKa = 2.97RR296 pKa = 11.84DD297 pKa = 3.98GASDD301 pKa = 3.68GGGVGAPQDD310 pKa = 3.69QEE312 pKa = 4.82AVDD315 pKa = 4.64LFPVSSDD322 pKa = 2.91RR323 pKa = 11.84GEE325 pKa = 3.94TGEE328 pKa = 4.35AGGSGGEE335 pKa = 4.13GQEE338 pKa = 4.79MIGTDD343 pKa = 3.57AGDD346 pKa = 4.28EE347 pKa = 4.15ISGTSADD354 pKa = 4.13DD355 pKa = 4.02TIAGQGGHH363 pKa = 7.09DD364 pKa = 3.91MLSAGGGNDD373 pKa = 3.88NISGSDD379 pKa = 3.63GNDD382 pKa = 3.41TISAGSGHH390 pKa = 7.07DD391 pKa = 3.57NVGGGLGNDD400 pKa = 4.38EE401 pKa = 3.72IWGAGGNDD409 pKa = 3.68TIGAGRR415 pKa = 11.84GNDD418 pKa = 3.59YY419 pKa = 11.5VNAGPGNDD427 pKa = 3.54VANGGQGDD435 pKa = 4.06DD436 pKa = 4.22MIFGEE441 pKa = 5.53DD442 pKa = 3.82GDD444 pKa = 4.34DD445 pKa = 3.39TTGAGFGDD453 pKa = 4.05DD454 pKa = 3.67TVAGGSGHH462 pKa = 7.39DD463 pKa = 3.79SLGGGTGQDD472 pKa = 3.63MIAGHH477 pKa = 6.85GGNDD481 pKa = 3.81AIGGGEE487 pKa = 4.27GNDD490 pKa = 4.14SISGGDD496 pKa = 3.59GNDD499 pKa = 3.32FLAGGGRR506 pKa = 11.84DD507 pKa = 3.48DD508 pKa = 4.57VIYY511 pKa = 10.42GGSGADD517 pKa = 3.87TINGGDD523 pKa = 4.25GNDD526 pKa = 3.46ALYY529 pKa = 10.69GGTGADD535 pKa = 2.89VFVWNEE541 pKa = 3.85YY542 pKa = 9.8EE543 pKa = 4.92AGATDD548 pKa = 4.35TIYY551 pKa = 10.97GFQDD555 pKa = 3.26GVDD558 pKa = 3.8TLRR561 pKa = 11.84LADD564 pKa = 3.66VSNAPGTGLAGKK576 pKa = 10.09LDD578 pKa = 4.06ALNITDD584 pKa = 3.56IVVDD588 pKa = 4.55GVAGVRR594 pKa = 11.84VGYY597 pKa = 9.55QGNGIEE603 pKa = 4.69IPGLSAAQITLDD615 pKa = 4.46DD616 pKa = 3.76IMFII620 pKa = 4.45
MM1 pKa = 7.22TVDD4 pKa = 3.67IYY6 pKa = 11.58APTSSDD12 pKa = 3.49SLSLTEE18 pKa = 4.29LDD20 pKa = 4.34LYY22 pKa = 10.76HH23 pKa = 7.01MIMDD27 pKa = 3.7YY28 pKa = 10.64RR29 pKa = 11.84AASGLPAIPLSQSLTTVAGRR49 pKa = 11.84HH50 pKa = 5.04TLDD53 pKa = 3.32SYY55 pKa = 12.0HH56 pKa = 6.93NIEE59 pKa = 4.19AAGVSLPSGYY69 pKa = 10.5SSPSWIHH76 pKa = 5.42SWSDD80 pKa = 2.94AYY82 pKa = 11.61YY83 pKa = 10.37DD84 pKa = 4.24GSDD87 pKa = 3.41SSVMRR92 pKa = 11.84DD93 pKa = 3.12APSRR97 pKa = 11.84LGTGYY102 pKa = 9.85PGGGYY107 pKa = 9.74EE108 pKa = 4.17ISVAGAQSIRR118 pKa = 11.84QTLDD122 pKa = 2.6MWIDD126 pKa = 3.88SPGHH130 pKa = 5.29NQLLVNLNSYY140 pKa = 11.34SDD142 pKa = 3.94DD143 pKa = 2.94TWNAIGVGVGFNAPVSGGSFTYY165 pKa = 10.83GNYY168 pKa = 8.37FHH170 pKa = 6.87VWFGKK175 pKa = 10.07VADD178 pKa = 4.05PAGPPVIDD186 pKa = 3.55GTNAGEE192 pKa = 4.47LIEE195 pKa = 4.26MTNFADD201 pKa = 3.99EE202 pKa = 4.14VRR204 pKa = 11.84DD205 pKa = 3.55RR206 pKa = 11.84GGNDD210 pKa = 3.04TVIGYY215 pKa = 9.49SGNDD219 pKa = 3.07TFYY222 pKa = 11.37VGGGANDD229 pKa = 3.28FRR231 pKa = 11.84GGDD234 pKa = 3.49GTDD237 pKa = 2.87RR238 pKa = 11.84AVIPATFAQVTATVDD253 pKa = 3.29GSVAQITLNGTTSTYY268 pKa = 10.72RR269 pKa = 11.84GIEE272 pKa = 3.77VFAFSDD278 pKa = 3.54GTTRR282 pKa = 11.84ALGQLNGGTSGGDD295 pKa = 2.97RR296 pKa = 11.84DD297 pKa = 3.98GASDD301 pKa = 3.68GGGVGAPQDD310 pKa = 3.69QEE312 pKa = 4.82AVDD315 pKa = 4.64LFPVSSDD322 pKa = 2.91RR323 pKa = 11.84GEE325 pKa = 3.94TGEE328 pKa = 4.35AGGSGGEE335 pKa = 4.13GQEE338 pKa = 4.79MIGTDD343 pKa = 3.57AGDD346 pKa = 4.28EE347 pKa = 4.15ISGTSADD354 pKa = 4.13DD355 pKa = 4.02TIAGQGGHH363 pKa = 7.09DD364 pKa = 3.91MLSAGGGNDD373 pKa = 3.88NISGSDD379 pKa = 3.63GNDD382 pKa = 3.41TISAGSGHH390 pKa = 7.07DD391 pKa = 3.57NVGGGLGNDD400 pKa = 4.38EE401 pKa = 3.72IWGAGGNDD409 pKa = 3.68TIGAGRR415 pKa = 11.84GNDD418 pKa = 3.59YY419 pKa = 11.5VNAGPGNDD427 pKa = 3.54VANGGQGDD435 pKa = 4.06DD436 pKa = 4.22MIFGEE441 pKa = 5.53DD442 pKa = 3.82GDD444 pKa = 4.34DD445 pKa = 3.39TTGAGFGDD453 pKa = 4.05DD454 pKa = 3.67TVAGGSGHH462 pKa = 7.39DD463 pKa = 3.79SLGGGTGQDD472 pKa = 3.63MIAGHH477 pKa = 6.85GGNDD481 pKa = 3.81AIGGGEE487 pKa = 4.27GNDD490 pKa = 4.14SISGGDD496 pKa = 3.59GNDD499 pKa = 3.32FLAGGGRR506 pKa = 11.84DD507 pKa = 3.48DD508 pKa = 4.57VIYY511 pKa = 10.42GGSGADD517 pKa = 3.87TINGGDD523 pKa = 4.25GNDD526 pKa = 3.46ALYY529 pKa = 10.69GGTGADD535 pKa = 2.89VFVWNEE541 pKa = 3.85YY542 pKa = 9.8EE543 pKa = 4.92AGATDD548 pKa = 4.35TIYY551 pKa = 10.97GFQDD555 pKa = 3.26GVDD558 pKa = 3.8TLRR561 pKa = 11.84LADD564 pKa = 3.66VSNAPGTGLAGKK576 pKa = 10.09LDD578 pKa = 4.06ALNITDD584 pKa = 3.56IVVDD588 pKa = 4.55GVAGVRR594 pKa = 11.84VGYY597 pKa = 9.55QGNGIEE603 pKa = 4.69IPGLSAAQITLDD615 pKa = 4.46DD616 pKa = 3.76IMFII620 pKa = 4.45
Molecular weight: 62.18 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W4HL11|W4HL11_9RHOB Uncharacterized protein OS=Roseivivax marinus OX=1379903 GN=ATO8_05176 PE=4 SV=1
MM1 pKa = 7.74RR2 pKa = 11.84AATHH6 pKa = 6.31IAWPAPRR13 pKa = 11.84RR14 pKa = 11.84SPWTPLRR21 pKa = 11.84ATAAALLLCLSLPVTGAGPASAQTIGAMSPHH52 pKa = 7.13PGAGVSDD59 pKa = 3.72QAVEE63 pKa = 3.9VSLRR67 pKa = 11.84PKK69 pKa = 10.44VRR71 pKa = 11.84DD72 pKa = 3.35DD73 pKa = 4.09RR74 pKa = 11.84LPKK77 pKa = 10.38ARR79 pKa = 11.84WGTSARR85 pKa = 11.84GQTWTRR91 pKa = 11.84AAVSALRR98 pKa = 11.84GPASGLTDD106 pKa = 4.02IVPGDD111 pKa = 3.53IDD113 pKa = 3.84TWCPAYY119 pKa = 10.21EE120 pKa = 4.24EE121 pKa = 4.2NNRR124 pKa = 11.84RR125 pKa = 11.84LRR127 pKa = 11.84EE128 pKa = 4.19AFWVSLVSALAKK140 pKa = 10.45HH141 pKa = 5.99EE142 pKa = 4.29STYY145 pKa = 10.92RR146 pKa = 11.84PTVVGGGGRR155 pKa = 11.84WHH157 pKa = 7.22GLLQILPSTARR168 pKa = 11.84LYY170 pKa = 10.95GCRR173 pKa = 11.84AGSGAALQSGAANLSCGLRR192 pKa = 11.84IMARR196 pKa = 11.84TVARR200 pKa = 11.84DD201 pKa = 3.67GVVSAGMRR209 pKa = 11.84GVAADD214 pKa = 3.82WGPFHH219 pKa = 7.42SSTKK223 pKa = 9.54RR224 pKa = 11.84SDD226 pKa = 3.29MIAYY230 pKa = 7.28TRR232 pKa = 11.84SQAFCKK238 pKa = 9.91SVPGKK243 pKa = 10.58RR244 pKa = 11.84PMARR248 pKa = 11.84PDD250 pKa = 3.14VDD252 pKa = 3.54EE253 pKa = 4.32TVPLYY258 pKa = 11.16AEE260 pKa = 4.56IPPRR264 pKa = 11.84DD265 pKa = 3.53APVFSTQGRR274 pKa = 11.84LAPP277 pKa = 4.36
MM1 pKa = 7.74RR2 pKa = 11.84AATHH6 pKa = 6.31IAWPAPRR13 pKa = 11.84RR14 pKa = 11.84SPWTPLRR21 pKa = 11.84ATAAALLLCLSLPVTGAGPASAQTIGAMSPHH52 pKa = 7.13PGAGVSDD59 pKa = 3.72QAVEE63 pKa = 3.9VSLRR67 pKa = 11.84PKK69 pKa = 10.44VRR71 pKa = 11.84DD72 pKa = 3.35DD73 pKa = 4.09RR74 pKa = 11.84LPKK77 pKa = 10.38ARR79 pKa = 11.84WGTSARR85 pKa = 11.84GQTWTRR91 pKa = 11.84AAVSALRR98 pKa = 11.84GPASGLTDD106 pKa = 4.02IVPGDD111 pKa = 3.53IDD113 pKa = 3.84TWCPAYY119 pKa = 10.21EE120 pKa = 4.24EE121 pKa = 4.2NNRR124 pKa = 11.84RR125 pKa = 11.84LRR127 pKa = 11.84EE128 pKa = 4.19AFWVSLVSALAKK140 pKa = 10.45HH141 pKa = 5.99EE142 pKa = 4.29STYY145 pKa = 10.92RR146 pKa = 11.84PTVVGGGGRR155 pKa = 11.84WHH157 pKa = 7.22GLLQILPSTARR168 pKa = 11.84LYY170 pKa = 10.95GCRR173 pKa = 11.84AGSGAALQSGAANLSCGLRR192 pKa = 11.84IMARR196 pKa = 11.84TVARR200 pKa = 11.84DD201 pKa = 3.67GVVSAGMRR209 pKa = 11.84GVAADD214 pKa = 3.82WGPFHH219 pKa = 7.42SSTKK223 pKa = 9.54RR224 pKa = 11.84SDD226 pKa = 3.29MIAYY230 pKa = 7.28TRR232 pKa = 11.84SQAFCKK238 pKa = 9.91SVPGKK243 pKa = 10.58RR244 pKa = 11.84PMARR248 pKa = 11.84PDD250 pKa = 3.14VDD252 pKa = 3.54EE253 pKa = 4.32TVPLYY258 pKa = 11.16AEE260 pKa = 4.56IPPRR264 pKa = 11.84DD265 pKa = 3.53APVFSTQGRR274 pKa = 11.84LAPP277 pKa = 4.36
Molecular weight: 29.4 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1352812 |
29 |
5131 |
317.3 |
34.21 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.077 ± 0.056 | 0.814 ± 0.014 |
6.556 ± 0.046 | 6.243 ± 0.037 |
3.52 ± 0.02 | 9.164 ± 0.058 |
1.949 ± 0.022 | 4.6 ± 0.028 |
2.375 ± 0.035 | 9.834 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.545 ± 0.022 | 2.263 ± 0.023 |
5.22 ± 0.028 | 2.805 ± 0.023 |
7.317 ± 0.053 | 5.045 ± 0.029 |
5.688 ± 0.044 | 7.519 ± 0.03 |
1.391 ± 0.017 | 2.074 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
