
Pseudoflavonifractor sp. AF19-9AC
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Oscillospiraceae; Pseudoflavonifractor; unclassified Pseudoflavonifractor
Average proteome isoelectric point is 6.0
Get precalculated fractions of proteins
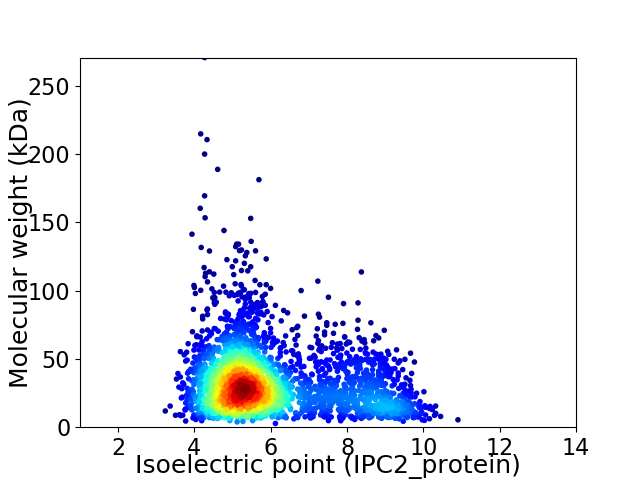
Virtual 2D-PAGE plot for 2880 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A416RC94|A0A416RC94_9FIRM DUF1349 domain-containing protein OS=Pseudoflavonifractor sp. AF19-9AC OX=2292244 GN=DWX58_06350 PE=4 SV=1
MM1 pKa = 7.49KK2 pKa = 8.69RR3 pKa = 11.84TKK5 pKa = 10.23YY6 pKa = 10.55RR7 pKa = 11.84SITVLLCALLVLSGCGARR25 pKa = 11.84TSEE28 pKa = 4.45EE29 pKa = 3.87TSSAQEE35 pKa = 4.01TSTSEE40 pKa = 3.69AAAVTTASTIDD51 pKa = 3.25TATIFSDD58 pKa = 3.45RR59 pKa = 11.84DD60 pKa = 3.26LEE62 pKa = 4.3GTYY65 pKa = 10.72DD66 pKa = 3.64EE67 pKa = 4.95SAAIAIQLSGDD78 pKa = 3.87SASCDD83 pKa = 3.16SDD85 pKa = 3.57AVTIEE90 pKa = 4.02GSRR93 pKa = 11.84ITIGAEE99 pKa = 3.73GVYY102 pKa = 10.37VLSGTLADD110 pKa = 4.01GQIVVNAGEE119 pKa = 4.08TDD121 pKa = 3.34KK122 pKa = 11.55VQLVLAGTDD131 pKa = 3.25ITSSTSAAIYY141 pKa = 10.4ALEE144 pKa = 4.12ADD146 pKa = 4.01KK147 pKa = 11.58VFVTLAEE154 pKa = 4.28GTEE157 pKa = 3.92NTLINGGEE165 pKa = 4.11YY166 pKa = 10.57VAIDD170 pKa = 4.58DD171 pKa = 4.38NNIDD175 pKa = 3.44AVIFAKK181 pKa = 10.09TDD183 pKa = 3.29LTLNGSGSLTVNAQAGHH200 pKa = 5.76GVVSKK205 pKa = 10.99DD206 pKa = 3.39DD207 pKa = 3.85LVITGGSYY215 pKa = 10.15TITAASHH222 pKa = 5.79GLSGKK227 pKa = 10.21DD228 pKa = 2.96VVAIADD234 pKa = 3.9GVFSITSGKK243 pKa = 10.48DD244 pKa = 3.0GIHH247 pKa = 6.93AEE249 pKa = 4.12NSDD252 pKa = 3.97DD253 pKa = 3.83LTLGTLYY260 pKa = 10.33IADD263 pKa = 3.88GSYY266 pKa = 10.68TITSQGDD273 pKa = 3.55TISAQGALQIDD284 pKa = 4.56GGTFDD289 pKa = 5.56LYY291 pKa = 10.65TGEE294 pKa = 4.72GSDD297 pKa = 3.9SVEE300 pKa = 4.19MTSGDD305 pKa = 3.17QFGQMGGGPRR315 pKa = 11.84GGIDD319 pKa = 3.24AQTVVTTTATEE330 pKa = 4.0EE331 pKa = 4.19DD332 pKa = 4.07TVSQKK337 pKa = 10.81GIKK340 pKa = 10.18GEE342 pKa = 4.04STYY345 pKa = 10.44TINGGVFTIDD355 pKa = 3.45SADD358 pKa = 3.71DD359 pKa = 3.6CLHH362 pKa = 6.73AGGVMTLAAGAFTLNSGDD380 pKa = 4.56DD381 pKa = 3.71AVHH384 pKa = 6.88CDD386 pKa = 4.34DD387 pKa = 4.82VLTIQSGTFTIPYY400 pKa = 8.75CYY402 pKa = 10.04EE403 pKa = 4.34GIEE406 pKa = 4.22GLSITIEE413 pKa = 4.58DD414 pKa = 3.85GTFDD418 pKa = 3.59ITSSDD423 pKa = 3.83DD424 pKa = 3.45GLNAAGGVDD433 pKa = 3.27SSGFGGQTQDD443 pKa = 2.96TFAADD448 pKa = 3.5SDD450 pKa = 4.26SFITINGGTLTIVSSGDD467 pKa = 3.71CVDD470 pKa = 3.84SNGALTINGGSLDD483 pKa = 3.92LTCNGSGNTTLDD495 pKa = 3.35CDD497 pKa = 3.75GTYY500 pKa = 11.0SNNGGDD506 pKa = 3.05ISTNDD511 pKa = 3.38GSEE514 pKa = 4.19DD515 pKa = 3.41NPGQMGGKK523 pKa = 10.16GGGQSGGGRR532 pKa = 11.84QDD534 pKa = 3.19GTNGGGRR541 pKa = 11.84PDD543 pKa = 3.63RR544 pKa = 11.84QTVPSASS551 pKa = 3.29
MM1 pKa = 7.49KK2 pKa = 8.69RR3 pKa = 11.84TKK5 pKa = 10.23YY6 pKa = 10.55RR7 pKa = 11.84SITVLLCALLVLSGCGARR25 pKa = 11.84TSEE28 pKa = 4.45EE29 pKa = 3.87TSSAQEE35 pKa = 4.01TSTSEE40 pKa = 3.69AAAVTTASTIDD51 pKa = 3.25TATIFSDD58 pKa = 3.45RR59 pKa = 11.84DD60 pKa = 3.26LEE62 pKa = 4.3GTYY65 pKa = 10.72DD66 pKa = 3.64EE67 pKa = 4.95SAAIAIQLSGDD78 pKa = 3.87SASCDD83 pKa = 3.16SDD85 pKa = 3.57AVTIEE90 pKa = 4.02GSRR93 pKa = 11.84ITIGAEE99 pKa = 3.73GVYY102 pKa = 10.37VLSGTLADD110 pKa = 4.01GQIVVNAGEE119 pKa = 4.08TDD121 pKa = 3.34KK122 pKa = 11.55VQLVLAGTDD131 pKa = 3.25ITSSTSAAIYY141 pKa = 10.4ALEE144 pKa = 4.12ADD146 pKa = 4.01KK147 pKa = 11.58VFVTLAEE154 pKa = 4.28GTEE157 pKa = 3.92NTLINGGEE165 pKa = 4.11YY166 pKa = 10.57VAIDD170 pKa = 4.58DD171 pKa = 4.38NNIDD175 pKa = 3.44AVIFAKK181 pKa = 10.09TDD183 pKa = 3.29LTLNGSGSLTVNAQAGHH200 pKa = 5.76GVVSKK205 pKa = 10.99DD206 pKa = 3.39DD207 pKa = 3.85LVITGGSYY215 pKa = 10.15TITAASHH222 pKa = 5.79GLSGKK227 pKa = 10.21DD228 pKa = 2.96VVAIADD234 pKa = 3.9GVFSITSGKK243 pKa = 10.48DD244 pKa = 3.0GIHH247 pKa = 6.93AEE249 pKa = 4.12NSDD252 pKa = 3.97DD253 pKa = 3.83LTLGTLYY260 pKa = 10.33IADD263 pKa = 3.88GSYY266 pKa = 10.68TITSQGDD273 pKa = 3.55TISAQGALQIDD284 pKa = 4.56GGTFDD289 pKa = 5.56LYY291 pKa = 10.65TGEE294 pKa = 4.72GSDD297 pKa = 3.9SVEE300 pKa = 4.19MTSGDD305 pKa = 3.17QFGQMGGGPRR315 pKa = 11.84GGIDD319 pKa = 3.24AQTVVTTTATEE330 pKa = 4.0EE331 pKa = 4.19DD332 pKa = 4.07TVSQKK337 pKa = 10.81GIKK340 pKa = 10.18GEE342 pKa = 4.04STYY345 pKa = 10.44TINGGVFTIDD355 pKa = 3.45SADD358 pKa = 3.71DD359 pKa = 3.6CLHH362 pKa = 6.73AGGVMTLAAGAFTLNSGDD380 pKa = 4.56DD381 pKa = 3.71AVHH384 pKa = 6.88CDD386 pKa = 4.34DD387 pKa = 4.82VLTIQSGTFTIPYY400 pKa = 8.75CYY402 pKa = 10.04EE403 pKa = 4.34GIEE406 pKa = 4.22GLSITIEE413 pKa = 4.58DD414 pKa = 3.85GTFDD418 pKa = 3.59ITSSDD423 pKa = 3.83DD424 pKa = 3.45GLNAAGGVDD433 pKa = 3.27SSGFGGQTQDD443 pKa = 2.96TFAADD448 pKa = 3.5SDD450 pKa = 4.26SFITINGGTLTIVSSGDD467 pKa = 3.71CVDD470 pKa = 3.84SNGALTINGGSLDD483 pKa = 3.92LTCNGSGNTTLDD495 pKa = 3.35CDD497 pKa = 3.75GTYY500 pKa = 11.0SNNGGDD506 pKa = 3.05ISTNDD511 pKa = 3.38GSEE514 pKa = 4.19DD515 pKa = 3.41NPGQMGGKK523 pKa = 10.16GGGQSGGGRR532 pKa = 11.84QDD534 pKa = 3.19GTNGGGRR541 pKa = 11.84PDD543 pKa = 3.63RR544 pKa = 11.84QTVPSASS551 pKa = 3.29
Molecular weight: 55.35 kDa
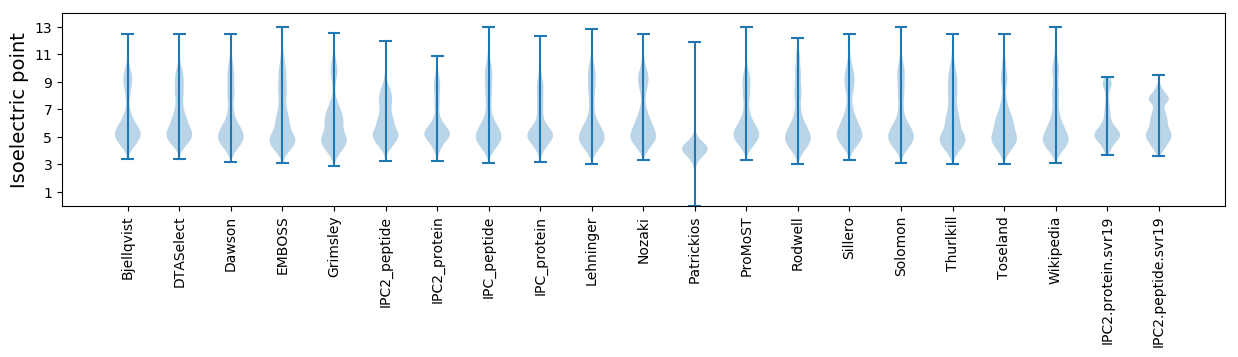
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A416REC9|A0A416REC9_9FIRM DNA polymerase III subunit delta OS=Pseudoflavonifractor sp. AF19-9AC OX=2292244 GN=DWX58_05520 PE=4 SV=1
MM1 pKa = 7.45LRR3 pKa = 11.84TYY5 pKa = 9.8QPKK8 pKa = 9.6KK9 pKa = 7.95RR10 pKa = 11.84QRR12 pKa = 11.84SKK14 pKa = 9.69EE15 pKa = 3.5HH16 pKa = 6.11GFRR19 pKa = 11.84KK20 pKa = 10.07RR21 pKa = 11.84MATRR25 pKa = 11.84NGRR28 pKa = 11.84KK29 pKa = 8.84VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.22GRR39 pKa = 11.84VRR41 pKa = 11.84LTHH44 pKa = 6.42
MM1 pKa = 7.45LRR3 pKa = 11.84TYY5 pKa = 9.8QPKK8 pKa = 9.6KK9 pKa = 7.95RR10 pKa = 11.84QRR12 pKa = 11.84SKK14 pKa = 9.69EE15 pKa = 3.5HH16 pKa = 6.11GFRR19 pKa = 11.84KK20 pKa = 10.07RR21 pKa = 11.84MATRR25 pKa = 11.84NGRR28 pKa = 11.84KK29 pKa = 8.84VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.22GRR39 pKa = 11.84VRR41 pKa = 11.84LTHH44 pKa = 6.42
Molecular weight: 5.42 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
892399 |
25 |
2581 |
309.9 |
34.23 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.286 ± 0.05 | 1.629 ± 0.022 |
5.433 ± 0.039 | 6.936 ± 0.058 |
3.721 ± 0.031 | 8.012 ± 0.051 |
1.796 ± 0.021 | 5.369 ± 0.045 |
4.534 ± 0.047 | 10.436 ± 0.06 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.775 ± 0.025 | 3.198 ± 0.032 |
4.286 ± 0.033 | 3.867 ± 0.027 |
5.588 ± 0.049 | 5.69 ± 0.045 |
5.583 ± 0.056 | 7.25 ± 0.038 |
1.174 ± 0.018 | 3.437 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
