
Rhizobium sp. BK696
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Rhizobiaceae; Rhizobium/Agrobacterium group; Rhizobium; unclassified Rhizobium
Average proteome isoelectric point is 6.41
Get precalculated fractions of proteins
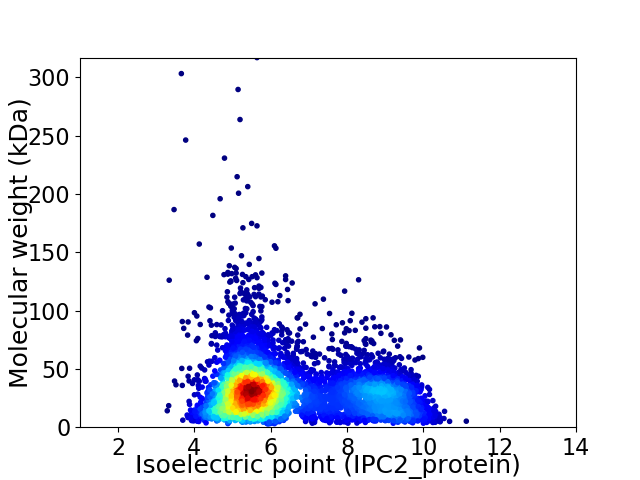
Virtual 2D-PAGE plot for 6267 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Q7SCN8|A0A4Q7SCN8_9RHIZ ATPase subunit of ABC transporter with duplicated ATPase domains OS=Rhizobium sp. BK696 OX=2512157 GN=EV569_4752 PE=3 SV=1
MM1 pKa = 6.43TTSIVNAVGRR11 pKa = 11.84PLYY14 pKa = 10.81YY15 pKa = 10.34SGSSTAWFSATKK27 pKa = 10.2AGPVQAGTAGNDD39 pKa = 3.82SMWGDD44 pKa = 3.83SAVKK48 pKa = 10.48VSMQGGTGDD57 pKa = 4.58DD58 pKa = 2.84IYY60 pKa = 11.73YY61 pKa = 10.51LYY63 pKa = 10.43SAKK66 pKa = 9.13NTAVEE71 pKa = 4.18QADD74 pKa = 3.95SGTDD78 pKa = 3.67TINTWMDD85 pKa = 3.03YY86 pKa = 9.34TLPDD90 pKa = 3.34NFEE93 pKa = 4.33NLTVTGNGRR102 pKa = 11.84YY103 pKa = 9.81AFGNNADD110 pKa = 4.47NIISGASGGQTIDD123 pKa = 3.13GGAGNDD129 pKa = 3.49VLIGGGGADD138 pKa = 2.77TFVFNHH144 pKa = 6.52GSGSDD149 pKa = 4.03LITDD153 pKa = 4.34FGADD157 pKa = 3.28DD158 pKa = 4.79VIRR161 pKa = 11.84MDD163 pKa = 4.05SFSSFADD170 pKa = 3.81LVAHH174 pKa = 6.03STQEE178 pKa = 4.17GADD181 pKa = 3.55VRR183 pKa = 11.84IDD185 pKa = 3.53TGNGEE190 pKa = 4.55SLVLANADD198 pKa = 3.18IDD200 pKa = 4.02QLSAAQFQLTLDD212 pKa = 3.81RR213 pKa = 11.84SHH215 pKa = 7.37LSLSFNDD222 pKa = 4.4DD223 pKa = 3.7FNSLSLRR230 pKa = 11.84DD231 pKa = 3.89GDD233 pKa = 4.56QGTWDD238 pKa = 3.83AKK240 pKa = 10.31FWWAPEE246 pKa = 3.93RR247 pKa = 11.84GGTLSGNGEE256 pKa = 4.23QQWYY260 pKa = 9.56INPSYY265 pKa = 10.62EE266 pKa = 4.16PTASASPFSVNDD278 pKa = 4.86GILTISANPASAALQQQIDD297 pKa = 4.4GYY299 pKa = 10.59DD300 pKa = 3.59YY301 pKa = 11.44TSGLLNTHH309 pKa = 6.12SSFSQTYY316 pKa = 8.43GYY318 pKa = 10.83FEE320 pKa = 4.72IRR322 pKa = 11.84ADD324 pKa = 3.65MPDD327 pKa = 3.83DD328 pKa = 3.62QGVWPAFWLLPEE340 pKa = 5.2DD341 pKa = 4.82GSWPPEE347 pKa = 3.78LDD349 pKa = 3.52VVEE352 pKa = 5.06MRR354 pKa = 11.84GQDD357 pKa = 3.55PNTVNVTVHH366 pKa = 6.41SDD368 pKa = 3.25DD369 pKa = 4.36GGSHH373 pKa = 7.29IIDD376 pKa = 3.56STPVKK381 pKa = 10.24VASTEE386 pKa = 3.97GFHH389 pKa = 7.13NYY391 pKa = 8.37GVLWDD396 pKa = 4.2EE397 pKa = 5.65DD398 pKa = 4.25HH399 pKa = 6.3ITWYY403 pKa = 10.65FDD405 pKa = 3.54DD406 pKa = 4.12VAVAQADD413 pKa = 4.36TPDD416 pKa = 4.22DD417 pKa = 3.78MHH419 pKa = 8.67DD420 pKa = 3.45PMYY423 pKa = 10.32MLVNLAVGGAAGSPADD439 pKa = 3.72GLAGGSEE446 pKa = 4.06MNIDD450 pKa = 3.96YY451 pKa = 10.73IRR453 pKa = 11.84AYY455 pKa = 9.74EE456 pKa = 4.08LADD459 pKa = 3.2NTVPPEE465 pKa = 3.98PAVEE469 pKa = 4.7FYY471 pKa = 11.19VV472 pKa = 3.62
MM1 pKa = 6.43TTSIVNAVGRR11 pKa = 11.84PLYY14 pKa = 10.81YY15 pKa = 10.34SGSSTAWFSATKK27 pKa = 10.2AGPVQAGTAGNDD39 pKa = 3.82SMWGDD44 pKa = 3.83SAVKK48 pKa = 10.48VSMQGGTGDD57 pKa = 4.58DD58 pKa = 2.84IYY60 pKa = 11.73YY61 pKa = 10.51LYY63 pKa = 10.43SAKK66 pKa = 9.13NTAVEE71 pKa = 4.18QADD74 pKa = 3.95SGTDD78 pKa = 3.67TINTWMDD85 pKa = 3.03YY86 pKa = 9.34TLPDD90 pKa = 3.34NFEE93 pKa = 4.33NLTVTGNGRR102 pKa = 11.84YY103 pKa = 9.81AFGNNADD110 pKa = 4.47NIISGASGGQTIDD123 pKa = 3.13GGAGNDD129 pKa = 3.49VLIGGGGADD138 pKa = 2.77TFVFNHH144 pKa = 6.52GSGSDD149 pKa = 4.03LITDD153 pKa = 4.34FGADD157 pKa = 3.28DD158 pKa = 4.79VIRR161 pKa = 11.84MDD163 pKa = 4.05SFSSFADD170 pKa = 3.81LVAHH174 pKa = 6.03STQEE178 pKa = 4.17GADD181 pKa = 3.55VRR183 pKa = 11.84IDD185 pKa = 3.53TGNGEE190 pKa = 4.55SLVLANADD198 pKa = 3.18IDD200 pKa = 4.02QLSAAQFQLTLDD212 pKa = 3.81RR213 pKa = 11.84SHH215 pKa = 7.37LSLSFNDD222 pKa = 4.4DD223 pKa = 3.7FNSLSLRR230 pKa = 11.84DD231 pKa = 3.89GDD233 pKa = 4.56QGTWDD238 pKa = 3.83AKK240 pKa = 10.31FWWAPEE246 pKa = 3.93RR247 pKa = 11.84GGTLSGNGEE256 pKa = 4.23QQWYY260 pKa = 9.56INPSYY265 pKa = 10.62EE266 pKa = 4.16PTASASPFSVNDD278 pKa = 4.86GILTISANPASAALQQQIDD297 pKa = 4.4GYY299 pKa = 10.59DD300 pKa = 3.59YY301 pKa = 11.44TSGLLNTHH309 pKa = 6.12SSFSQTYY316 pKa = 8.43GYY318 pKa = 10.83FEE320 pKa = 4.72IRR322 pKa = 11.84ADD324 pKa = 3.65MPDD327 pKa = 3.83DD328 pKa = 3.62QGVWPAFWLLPEE340 pKa = 5.2DD341 pKa = 4.82GSWPPEE347 pKa = 3.78LDD349 pKa = 3.52VVEE352 pKa = 5.06MRR354 pKa = 11.84GQDD357 pKa = 3.55PNTVNVTVHH366 pKa = 6.41SDD368 pKa = 3.25DD369 pKa = 4.36GGSHH373 pKa = 7.29IIDD376 pKa = 3.56STPVKK381 pKa = 10.24VASTEE386 pKa = 3.97GFHH389 pKa = 7.13NYY391 pKa = 8.37GVLWDD396 pKa = 4.2EE397 pKa = 5.65DD398 pKa = 4.25HH399 pKa = 6.3ITWYY403 pKa = 10.65FDD405 pKa = 3.54DD406 pKa = 4.12VAVAQADD413 pKa = 4.36TPDD416 pKa = 4.22DD417 pKa = 3.78MHH419 pKa = 8.67DD420 pKa = 3.45PMYY423 pKa = 10.32MLVNLAVGGAAGSPADD439 pKa = 3.72GLAGGSEE446 pKa = 4.06MNIDD450 pKa = 3.96YY451 pKa = 10.73IRR453 pKa = 11.84AYY455 pKa = 9.74EE456 pKa = 4.08LADD459 pKa = 3.2NTVPPEE465 pKa = 3.98PAVEE469 pKa = 4.7FYY471 pKa = 11.19VV472 pKa = 3.62
Molecular weight: 50.4 kDa
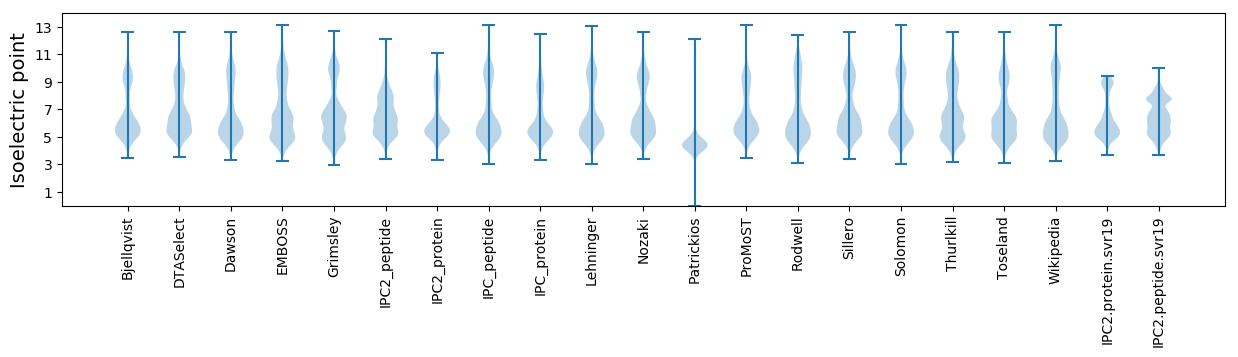
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Q7T0M4|A0A4Q7T0M4_9RHIZ Uncharacterized protein OS=Rhizobium sp. BK696 OX=2512157 GN=EV569_0170 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.27GGQKK29 pKa = 9.87VLSARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.03RR41 pKa = 11.84LSAA44 pKa = 4.03
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.27GGQKK29 pKa = 9.87VLSARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.03RR41 pKa = 11.84LSAA44 pKa = 4.03
Molecular weight: 5.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1901983 |
29 |
2960 |
303.5 |
32.99 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.003 ± 0.037 | 0.777 ± 0.01 |
5.758 ± 0.026 | 5.748 ± 0.03 |
3.948 ± 0.02 | 8.304 ± 0.034 |
1.989 ± 0.015 | 5.798 ± 0.025 |
3.891 ± 0.024 | 9.848 ± 0.034 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.585 ± 0.016 | 2.888 ± 0.019 |
4.802 ± 0.02 | 3.144 ± 0.02 |
6.428 ± 0.032 | 5.937 ± 0.026 |
5.32 ± 0.025 | 7.222 ± 0.026 |
1.267 ± 0.013 | 2.343 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
